4 Analysis
I move on to the analyses.
We have about a dozen models to run.
All of these models should take into account that the data are nested (in participants and days).
That means quite complex model structures, especially when we have many predictors.
The lme4 package can handle such multi-level models, but in my experience suffers from many convergence issues that a Bayesian approach (i.e., the brms package) can handle better (see this paper for a comparison on convergence).
Moreover, corrections for multiple comparisons are less of a problem with Bayesian models.
Last, although the models here are mostly exploratory, we can still incorporate prior knowledge from the literature (and common sense about our variables).
4.1 Research questions
We have a total of 9 research questions. I divide them in three blocks:
- To what extent do specific person-level variables such as personality and motivational factors shape the accuracy of social media time engagement?
- To what extent do subjective experiences such as mood predict and or interact with person-level factors to shape the accuracy of social media time engagement?
- What are the unique relations relating objective and subjective engagement to well-being outcomes?
Each of those questions comes with a number of models. I’ll structure the analysis section according to those blocks.
4.2 Data preparation
I processed the dat data set in previous sections.
Specifying priors on the untransformed variables can be tough.
In some analyses (e.g., predicting social media use) I’ll use centered predictors to make it easier to interpret the intercept.
In other analyses (e.g., predicting error), I want to standardize outcomes and predictors of interest because standardized variables make it easier to choose sensible priors, especially because we can use standardized effect sizes reported in the literature.
Standardizing also makes it easier to interpret the effect sizes.
First, I transform all variables of interest (into new variables).
dat <-
dat %>%
# center personality variables
mutate(
across(
c(
autonomy_state:relatedness_state,
satisfied:enjoyable,
autonomy_trait:openness
),
~ scale(.x, center = TRUE, scale = FALSE),
.names = "{col}_c" # add "_c" suffix to new variables for "centered"
)
)NAs there are per variable.
Table 4.1 shows that missing values aren’t a big problem.
The missings come from rows where participants didn’t answer the diary, but did report screen time mostly.
| name | value |
|---|---|
| social_media_subjective | 51 |
| pickups_subjective | 38 |
| notifications_subjective | 38 |
| social_media_objective | 51 |
| error | 53 |
| pickups_objective | 30 |
| weekly_notifications | 0 |
| well_being_state | 40 |
| autonomy_state | 39 |
| competence_state | 39 |
| relatedness_state | 39 |
| satisfied | 42 |
| boring | 42 |
| stressful | 41 |
| enjoyable | 44 |
| autonomy_trait | 0 |
| competence_trait | 0 |
| relatedness_trait | 0 |
| extraversion | 0 |
| agreeableness | 0 |
| conscientiousness | 0 |
| neuroticism | 0 |
| openness | 0 |
4.3 Do trait variables predict social media use and accuracy?
Our first research question asks whether personality traits and trait motivations predict the accuracy of social media time engagement. Specifically, we have three models that address the following sub-questions:
- Do person-level variables predict objective-only engagement?
- Do person-level variables predict subjective-only engagement?
- Do person-level variables predict accuracy?
I’ll construct a model for each of those three questions.
Our dependent variables are clear-cut: social_media_objective, social_media_subjective, and error.
Our predictors will be variables on the person-level, so trait/personality variables: the Big Five and the three self-determination motivations.
Note, I saved all brms model objects and load them below.
Each model takes quite some time and the files get large (a total of half a GB).
If you’re reproducing the analysis and don’t want to run the models yourself, you can download the model objects from the OSF.
For that, you need to set the code chunk below to eval=TRUE, which creates a model/ directory with all models inside.
# create directory
dir.create("models/", FALSE, TRUE)
# download models
osf_retrieve_node("https://osf.io/7byvt/") %>%
osf_ls_nodes() %>%
filter(name == "models") %>%
osf_ls_files(
.,
n_max = Inf
) %>%
osf_download(
.,
path = here("models"),
progress = FALSE
)4.3.1 Model 1: Trait variables predicting objective use
First, I choose sensible priors. There’s some literature out there on Big Five and smartphone use, as well as motivations and smartphone use. If we only knew the mean and variance of the social media estimates, a Gaussian distribution would be most appropriate. However, we do know more than just those two parameters. Namely, we know that the scale is continuous (i.e., time) and cannot be less than zero. Also, if we look at the distribution of time on an activity, the variance usually increases with the mean. Therefore, a gamma distribution appears more adequate to me.
That means the models will use a log-link, which makes it hard to have an intuition about prior distributions (at least for me). Thus, I follow the recommendations of McElreath and simulate the priors.
- For the intercept, we can look at previous research. For example, this paper has average phone use times of about two hours per participant per day. Here, we only looked at social media use, so as a guess-timate with reasonable uncertainty, I’ll choose a lognormal distribution with a meanlog of 4.5 and a meansd of 0.8. See the upper left panel of Figure 4.1. That intercept has mots its value below two hours, but allows substantial skew for a couple of heavy users.
- For the shape of the gamma distribution, I also played around and settled on one that somewhat resembles our assumptions on the intercept, such that the majority of values will be below five hours with a couple of heavy users (see left side upper panel).
- For the Big Five fixed effects, this paper predicted social media use with the Big Five over two time period in a longitudinal design. They found that only neuroticism was related to social media use, but that effect was extremely small (\(\beta\) = .028). None of the other effects were large. Therefore, I’ll use rather flat, weakly regularizing priors for those effects. In 4.1, lower panel, I’ll use the prior on the left, because it’s skeptical, but regularizing. The one on the right might be too optimistic by suggesting a small positive effect.
- For basic psychological needs, there’s quite some literature on these needs and pathological smartphone use (“smartphone addiction”). However, there’s little info we could use for priors, so I’ll just go with the same prior as for the Big Five. See the top right of the Figure below.
- For all other parameters, I’ll take the
brmsdefault priors.
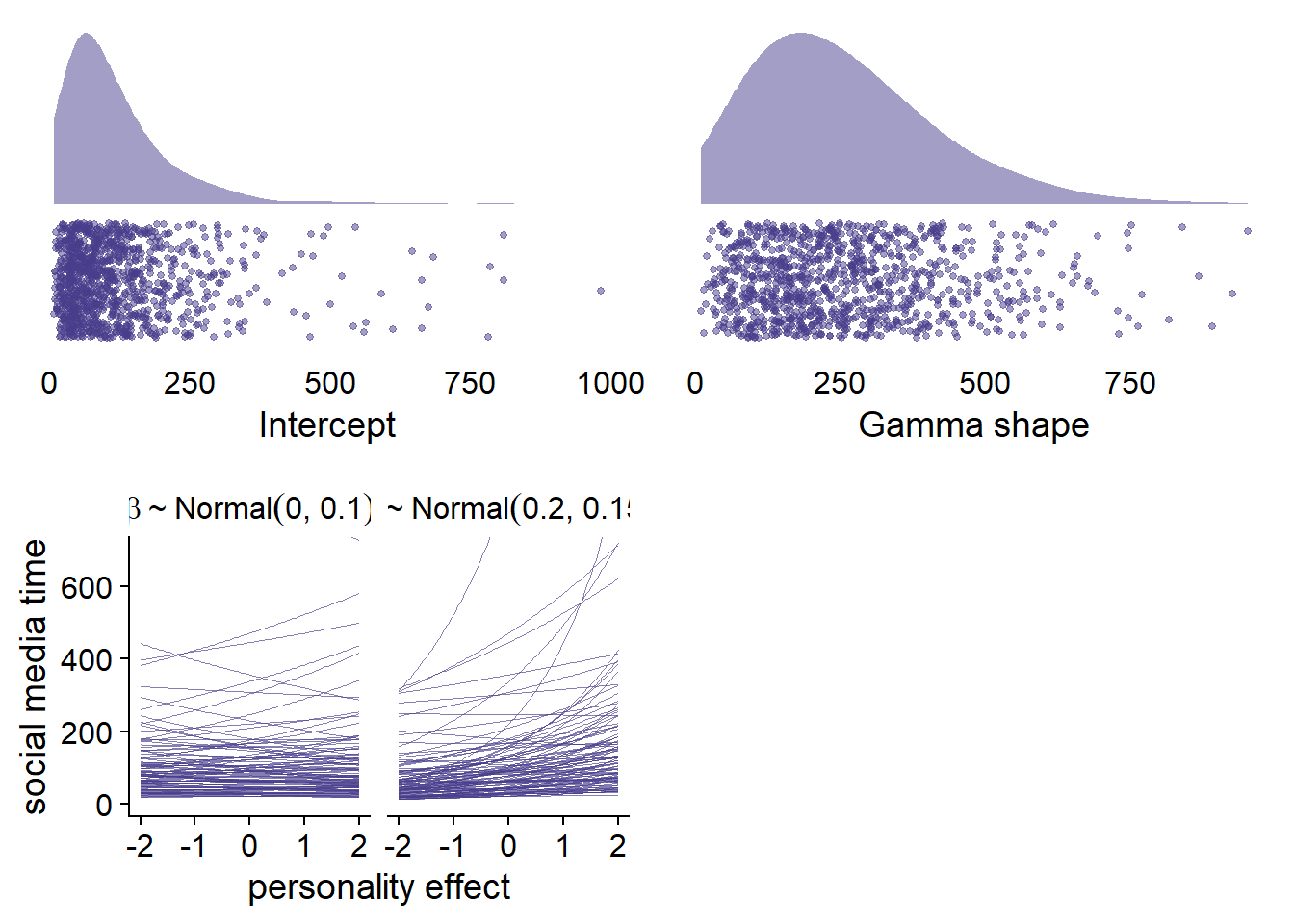
Figure 4.1: Prior simmulations for Model 1
Let’s set those priors we simulated above.
priors_model1 <-
c(
# intercept
prior(normal(4.5, 0.8), class = Intercept),
# prior on effects
prior(normal(0, 0.1), class = b),
# all other effects
prior(gamma(2.5, 100), class = shape)
)Alright, time to run the model. Luckily, none of these variables have missing values, so I won’t need to model missings in this model. Note that I ran the block below once and stored the model. Those fit objects are too large for Github, so you can download them from the OSF. See the instructions in the Readme of the project.
model1 <-
brm(
data = dat,
family = Gamma(link = "log"),
prior = priors_model1,
social_media_objective ~
1 +
openness_c +
conscientiousness_c +
extraversion_c +
agreeableness_c +
neuroticism_c +
competence_trait_c +
relatedness_trait_c +
autonomy_trait_c +
(1 | id) +
(1 | day),
iter = 5000,
warmup = 2000,
chains = 4,
cores = 4,
seed = 42,
control = list(adapt_delta = 0.99),
file = here("models", "model1")
)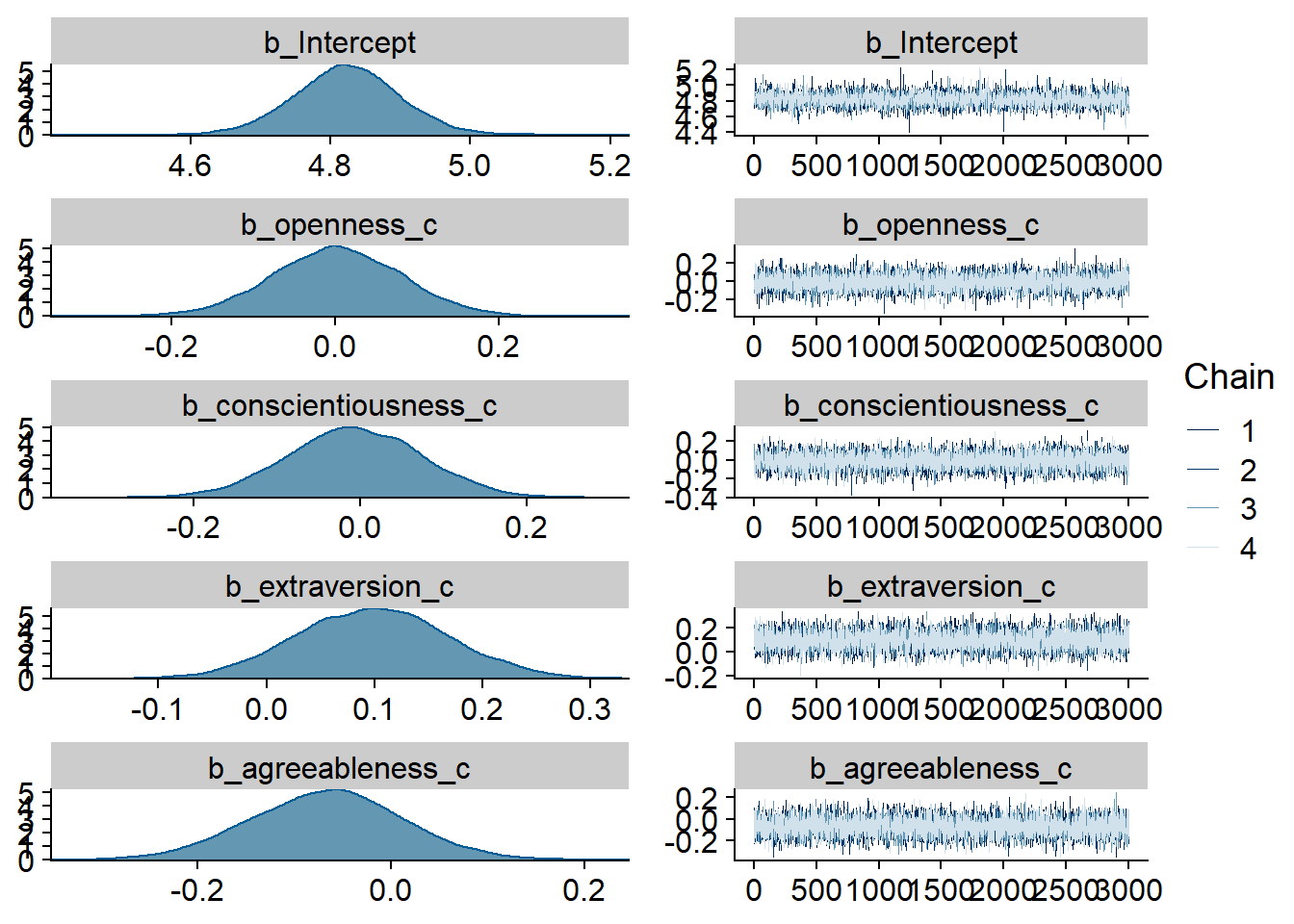
Figure 4.2: Traceplots and posterior distributions for Model 1
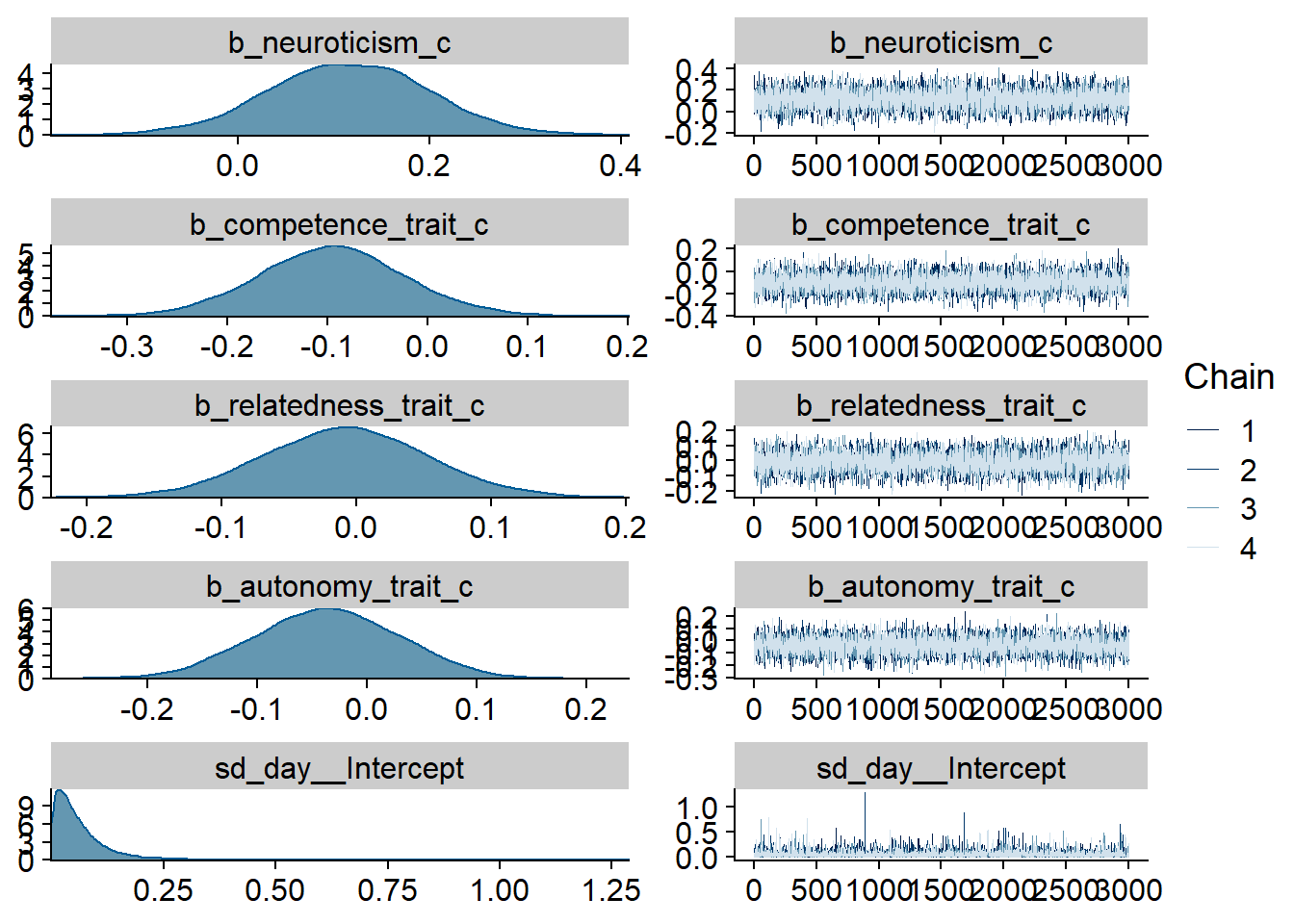
Figure 4.3: Traceplots and posterior distributions for Model 1
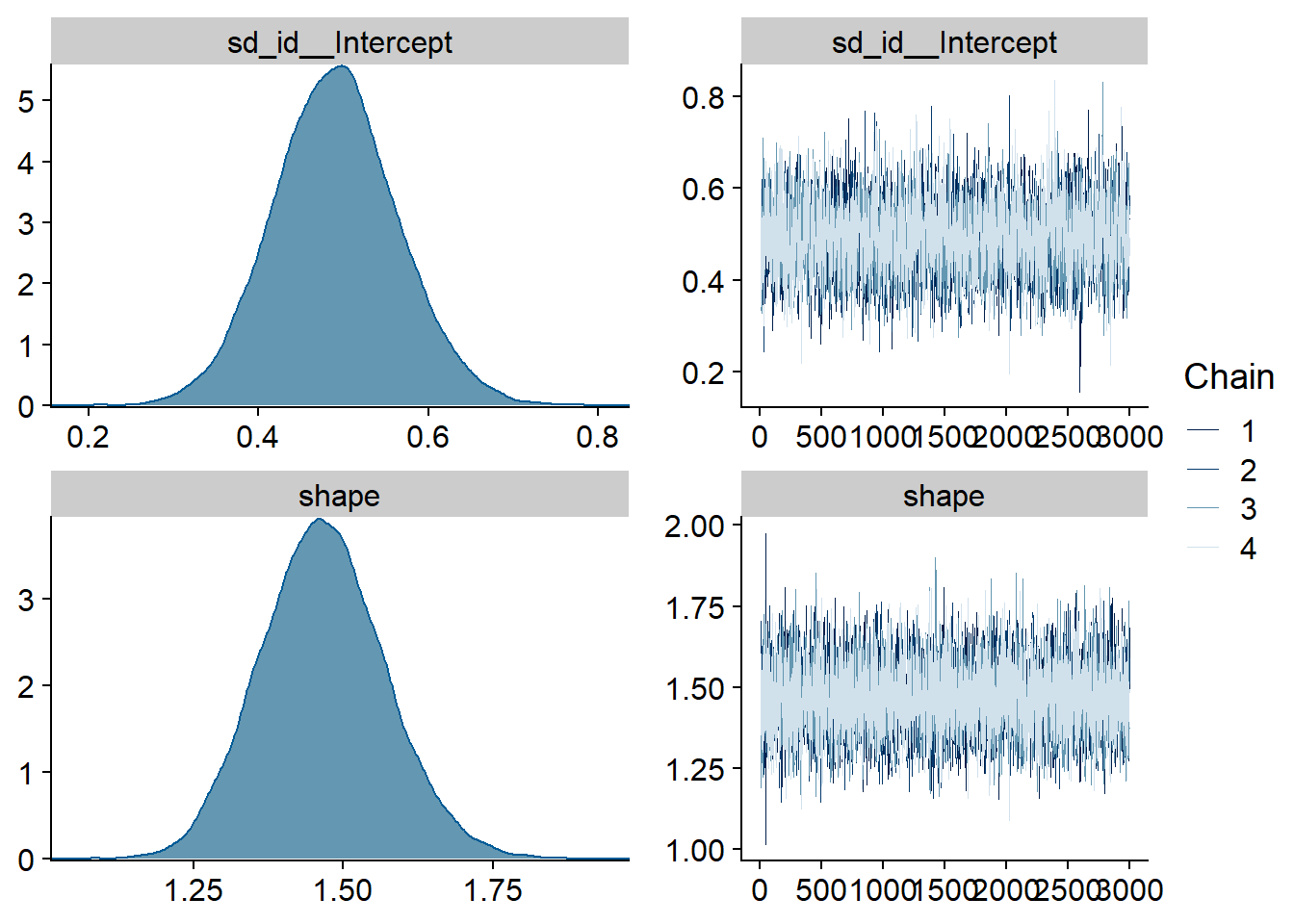
Figure 4.4: Traceplots and posterior distributions for Model 1
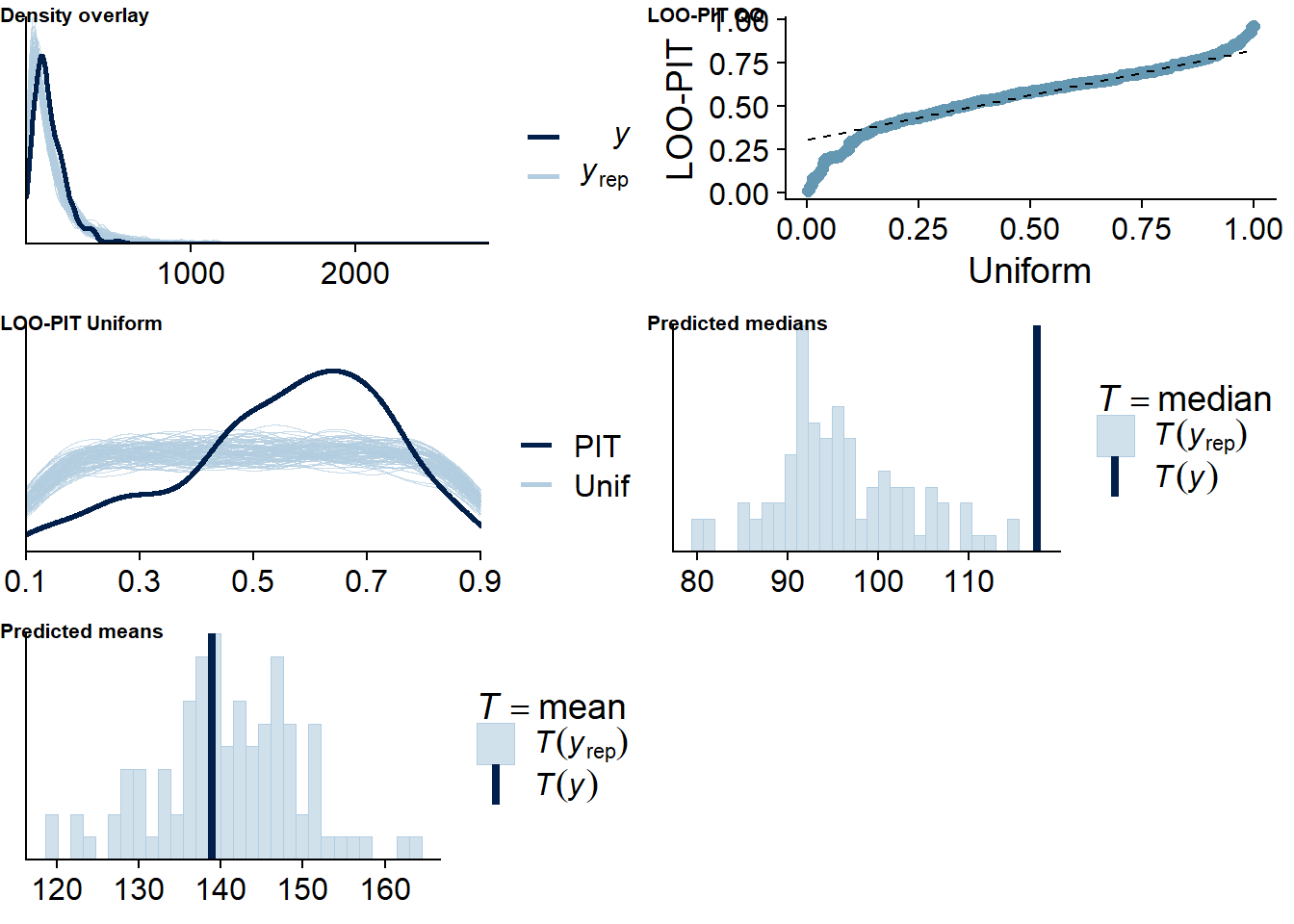
Figure 4.5: Posterior predictive checks for Model 1
Let’s also check for potentially influential values. None are tagged as influential, which increases my trust in the model.
##
## Computed from 12000 by 428 log-likelihood matrix
##
## Estimate SE
## elpd_loo -2440.0 14.4
## p_loo 20.7 1.4
## looic 4880.0 28.8
## ------
## Monte Carlo SE of elpd_loo is 0.1.
##
## All Pareto k estimates are good (k < 0.5).
## See help('pareto-k-diagnostic') for details.Alright, time to look at the summary: neuroticism and competence are the only predictors whose posterior distribution isn’t centered on zero. That said, we cannot be 95% certain the true value doesn’t contain zero.
## Family: gamma
## Links: mu = log; shape = identity
## Formula: social_media_objective ~ 1 + openness_c + conscientiousness_c + extraversion_c + agreeableness_c + neuroticism_c + competence_trait_c + relatedness_trait_c + autonomy_trait_c + (1 | id) + (1 | day)
## Data: study1 (Number of observations: 428)
## Samples: 4 chains, each with iter = 5000; warmup = 2000; thin = 1;
## total post-warmup samples = 12000
##
## Priors:
## b ~ normal(0, 0.1)
## Intercept ~ normal(4.5, 0.8)
## sd ~ student_t(3, 0, 2.5)
## shape ~ gamma(2.5, 100)
##
## Group-Level Effects:
## ~day (Number of levels: 5)
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## sd(Intercept) 0.06 0.07 0.00 0.23 1.00 5873 6154
##
## ~id (Number of levels: 94)
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## sd(Intercept) 0.49 0.07 0.35 0.64 1.00 3739 6583
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## Intercept 4.82 0.08 4.67 4.97 1.00 5813 6625
## openness_c 0.00 0.08 -0.16 0.16 1.00 11686 9877
## conscientiousness_c -0.01 0.08 -0.16 0.15 1.00 12552 9686
## extraversion_c 0.10 0.07 -0.04 0.24 1.00 9902 9429
## agreeableness_c -0.07 0.08 -0.22 0.09 1.00 10920 9571
## neuroticism_c 0.12 0.08 -0.05 0.28 1.00 11221 9205
## competence_trait_c -0.10 0.07 -0.24 0.05 1.00 8796 7816
## relatedness_trait_c -0.01 0.06 -0.13 0.11 1.00 9211 9015
## autonomy_trait_c -0.04 0.07 -0.17 0.09 1.00 8964 8810
##
## Family Specific Parameters:
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## shape 1.47 0.10 1.27 1.68 1.00 9250 8357
##
## Samples were drawn using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).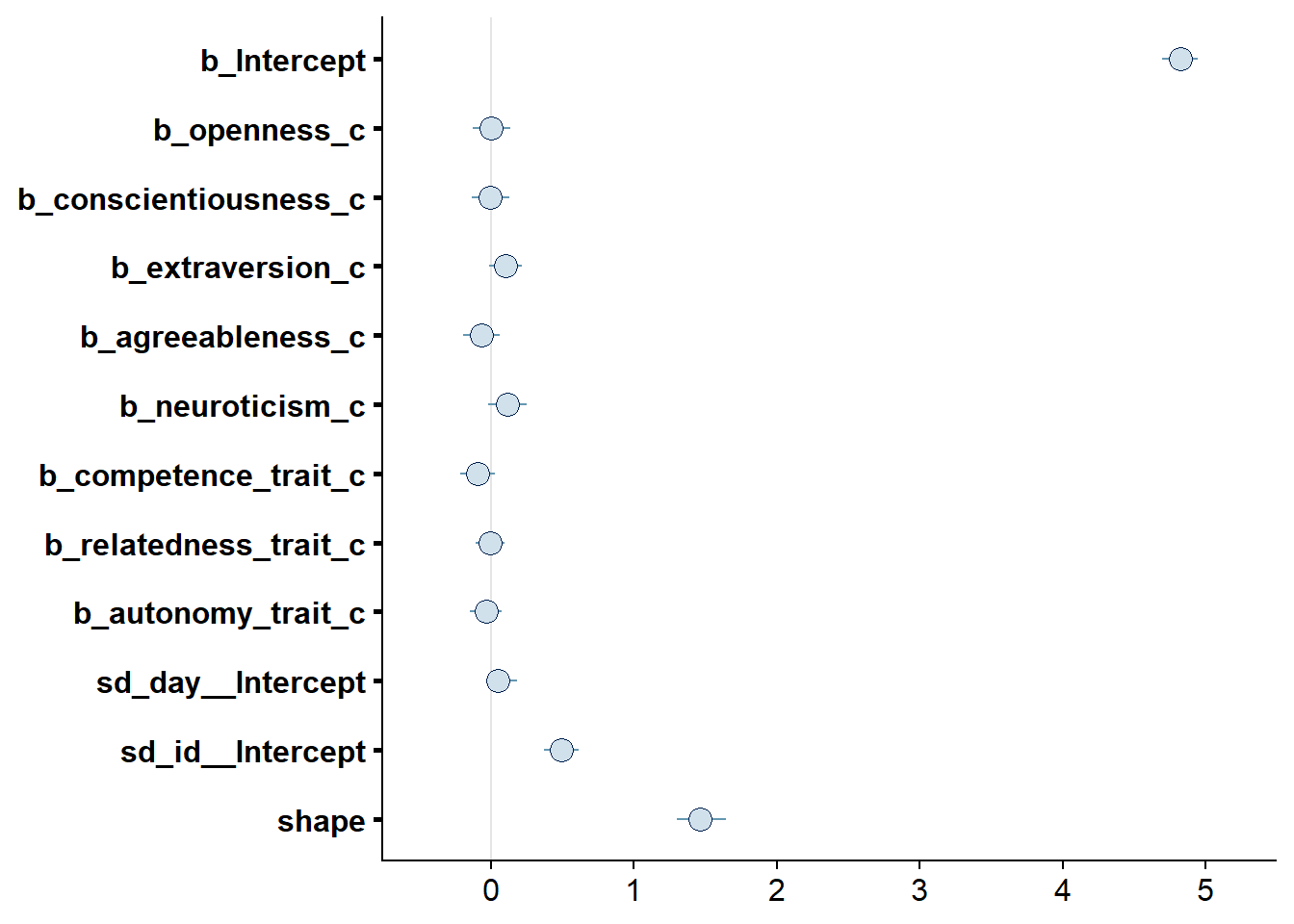
Figure 4.6: Effects plot for Model 1
4.3.2 Model 2: Trait variables predicting subjective use
Next up, I predict subjective use from the same predictors. We know that objective and subjective use aren’t perfectly correlated. Then again, the priors for Model 1 were only weakly regularizing, which is why I use the same priors again.
priors_model2 <-
c(
# intercept
prior(normal(4.5, 0.8), class = Intercept),
# prior on effects
prior(normal(0, 0.1), class = b),
# all other effects
prior(gamma(2.5, 100), class = shape)
)Alright, time to run the model. The subjective social media estimate had one missing value. Usually I’d impute missing values during model fitting, but with one, I think it’s safe to drop it.
model2 <-
brm(
data = dat,
family = Gamma(link = "log"),
prior = priors_model2,
social_media_subjective ~
1 +
openness_c +
conscientiousness_c +
extraversion_c +
agreeableness_c +
neuroticism_c +
competence_trait_c +
relatedness_trait_c +
autonomy_trait_c +
(1 | id) +
(1 | day),
iter = 5000,
warmup = 2000,
chains = 4,
cores = 4,
seed = 42,
control = list(adapt_delta = 0.999),
file = here("models", "model2")
)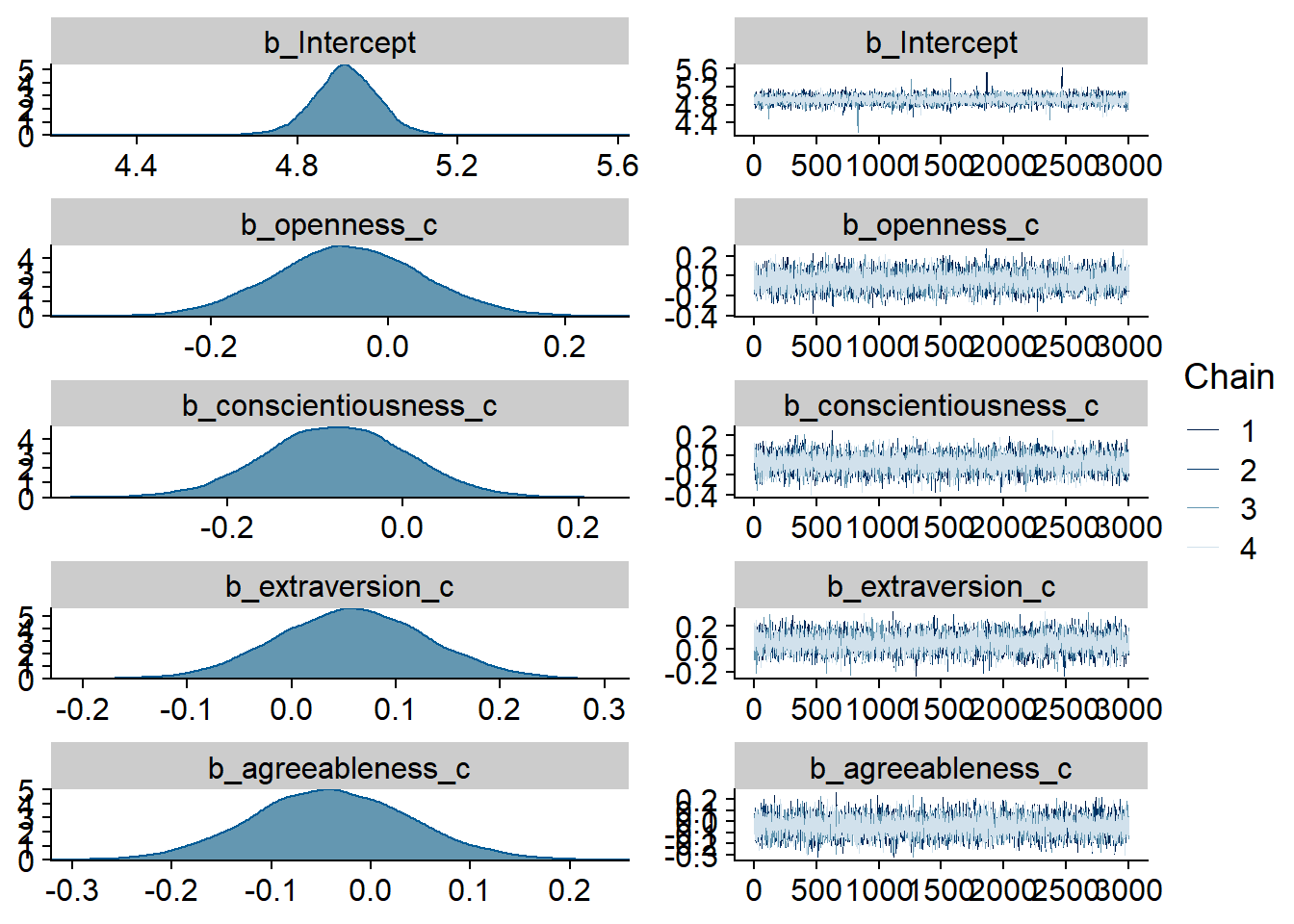
Figure 4.7: Traceplots and posterior distributions for Model 2
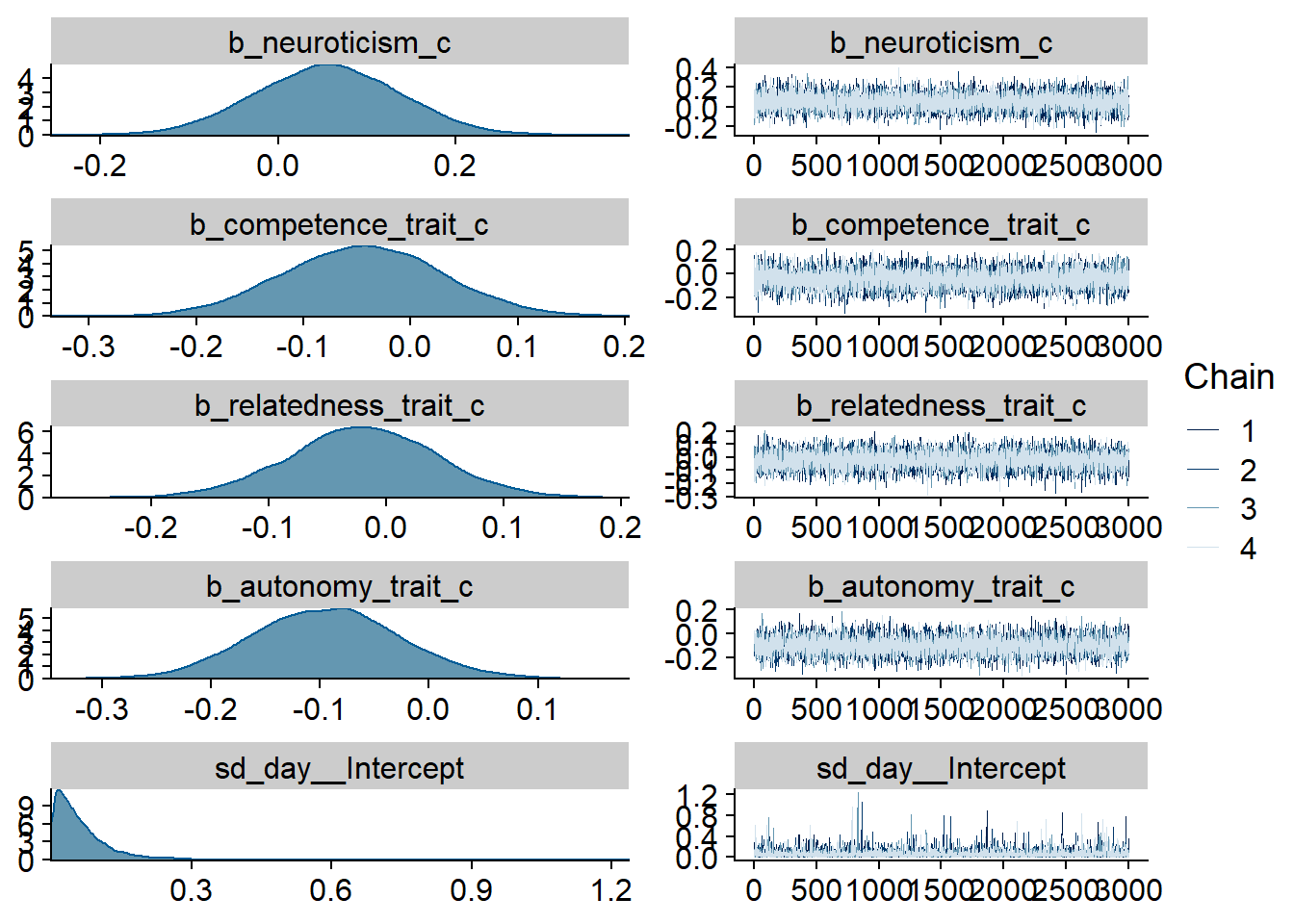
Figure 4.8: Traceplots and posterior distributions for Model 2
Figure 4.9: Traceplots and posterior distributions for Model 2
Figure 4.10: Posterior predictive checks for Model 2
Let’s again check for potentially influential values. The model diagnostics look good. Even though there were several rather large raw values on the outcome variable, the model expects them because we model the outcome as a Gamma distribution. We need to calculate ELPD directly, which shows no outliers.
## 1 problematic observation(s) found.
## The model will be refit 1 times.##
## Fitting model 1 out of 1 (leaving out observation 392)## Start sampling##
## Computed from 12000 by 428 log-likelihood matrix
##
## Estimate SE
## elpd_loo -2502.2 15.9
## p_loo 24.6 1.6
## looic 5004.4 31.8
## ------
## Monte Carlo SE of elpd_loo is 0.1.
##
## Pareto k diagnostic values:
## Count Pct. Min. n_eff
## (-Inf, 0.5] (good) 421 98.4% 412
## (0.5, 0.7] (ok) 7 1.6% 2552
## (0.7, 1] (bad) 0 0.0% <NA>
## (1, Inf) (very bad) 0 0.0% <NA>
##
## All Pareto k estimates are ok (k < 0.7).
## See help('pareto-k-diagnostic') for details.Alright, time to look at the summary. This time, all posterior distributions are mostly centered around zero, so we can be 95% (always conditional on the model) certain that personality traits and motivations are not meaningfully (i.e., large effect) related to self-reported social media use. Maybe autonomy gets close, but the posterior distributions still includes zero and small negative effects.
## Family: gamma
## Links: mu = log; shape = identity
## Formula: social_media_subjective ~ 1 + openness_c + conscientiousness_c + extraversion_c + agreeableness_c + neuroticism_c + competence_trait_c + relatedness_trait_c + autonomy_trait_c + (1 | id) + (1 | day)
## Data: study1 (Number of observations: 428)
## Samples: 4 chains, each with iter = 5000; warmup = 2000; thin = 1;
## total post-warmup samples = 12000
##
## Priors:
## b ~ normal(0, 0.1)
## Intercept ~ normal(4.5, 0.8)
## sd ~ student_t(3, 0, 2.5)
## shape ~ gamma(2.5, 100)
##
## Group-Level Effects:
## ~day (Number of levels: 5)
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## sd(Intercept) 0.07 0.07 0.00 0.25 1.00 5313 5290
##
## ~id (Number of levels: 94)
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## sd(Intercept) 0.51 0.08 0.36 0.66 1.00 3414 5977
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## Intercept 4.92 0.08 4.76 5.08 1.00 5160 5656
## openness_c -0.04 0.08 -0.21 0.12 1.00 9524 8983
## conscientiousness_c -0.08 0.08 -0.24 0.08 1.00 8766 9443
## extraversion_c 0.06 0.07 -0.08 0.20 1.00 7976 7997
## agreeableness_c -0.04 0.08 -0.19 0.12 1.00 8628 8552
## neuroticism_c 0.06 0.08 -0.10 0.21 1.00 9739 9263
## competence_trait_c -0.04 0.07 -0.19 0.10 1.00 8340 8625
## relatedness_trait_c -0.02 0.06 -0.14 0.10 1.00 7666 7707
## autonomy_trait_c -0.09 0.07 -0.22 0.04 1.00 7668 8591
##
## Family Specific Parameters:
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## shape 1.33 0.09 1.16 1.52 1.00 6185 9161
##
## Samples were drawn using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).
Figure 4.11: Effects plot for Model 2
4.3.3 Model 3: Trait variables predicting accuracy
For this model, it’s hard to choose informed priors, mainly because there’s no literature on the relation between personality traits and accuracy. Also, it’s unclear what distribution for accuracy we should expect prior to having seen the data. The few papers out there show that people generally overestimate their (social) media use, which would speak for centering the distribution on a positive value (i.e., positive error). Other than that, I’d expect a normal distribution, quite likely with fat tails, and possibly right skewed. Therefore, I’ll use a student-t distribution outcome family. I start with a t distribution as prior for the intercept that is slightly centered on overestimates (i.e., 20%), but with quite some potential for extreme values and fairly fat tails. See Figure 4.12 left panel, for a visualization.
As for the effects of the personality traits, I’ll be skeptical of any effects and thus use weakly regularizing normal priors that are centered on zero, with a small range for the effect: If a person goes from average on a trait to one above average, we’d expect that 95% of the effects should be between -50% and +50% (i.e., SD of 25). See the right panel in the figure below for a visualization.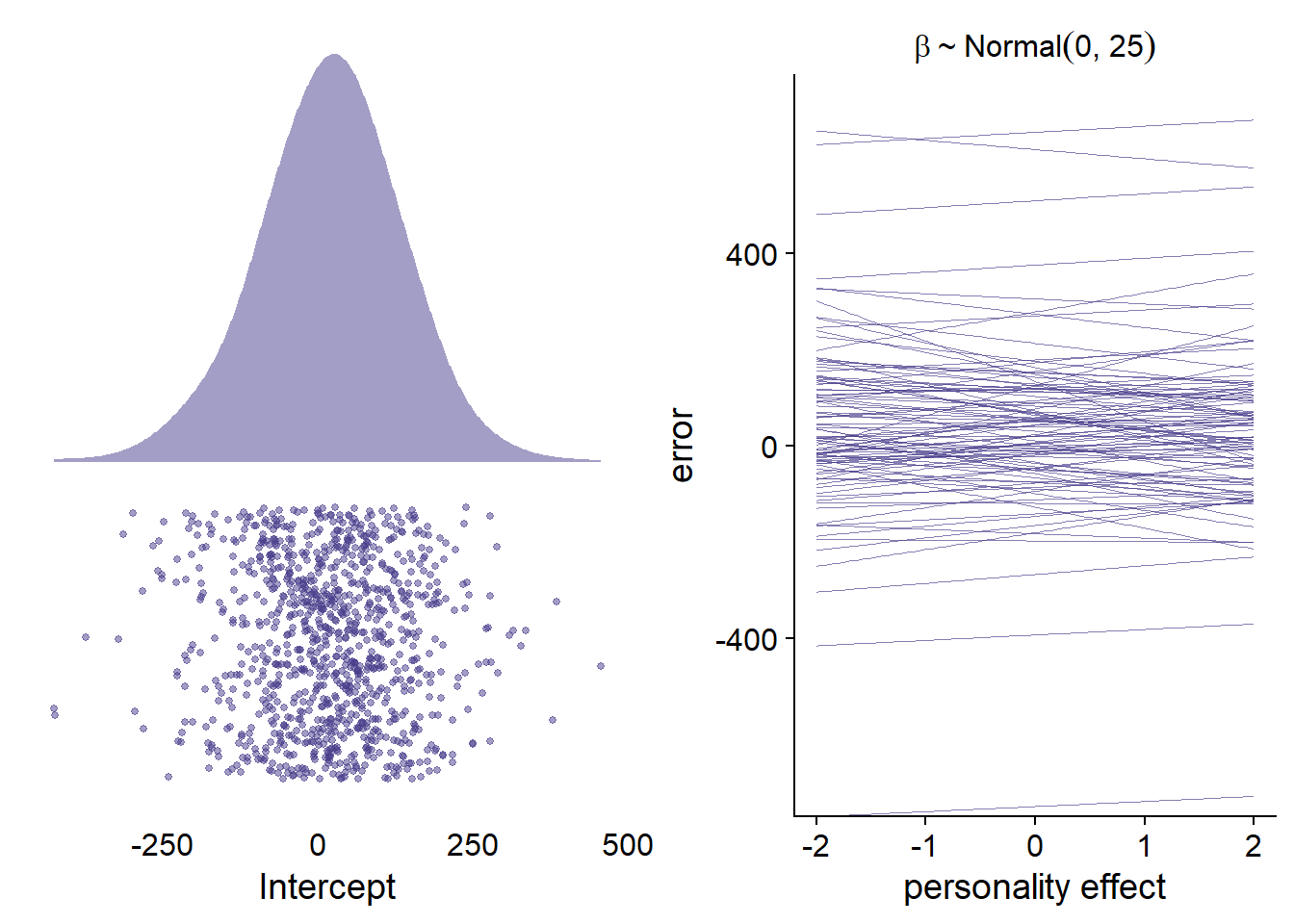
Figure 4.12: Prior simmulations for Model 2
Then let’s set those priors we simulated above.
priors_model3 <-
c(
# intercept
prior(student_t(10, 20, 100), class = Intercept),
# all other effects
prior(normal(0, 25), class = b)
)error had 53 missing values.
Again, with such few cases, I’m fine with dropping those during model fitting.
model3 <-
brm(
data = dat,
family = student,
prior = priors_model3,
error ~
1 +
openness_c +
conscientiousness_c +
extraversion_c +
agreeableness_c +
neuroticism_c +
competence_trait_c +
relatedness_trait_c +
autonomy_trait_c +
(1 | id) +
(1 | day),
iter = 5000,
warmup = 2000,
chains = 4,
cores = 4,
seed = 42,
control = list(adapt_delta = 0.99),
file = here("models", "model3")
)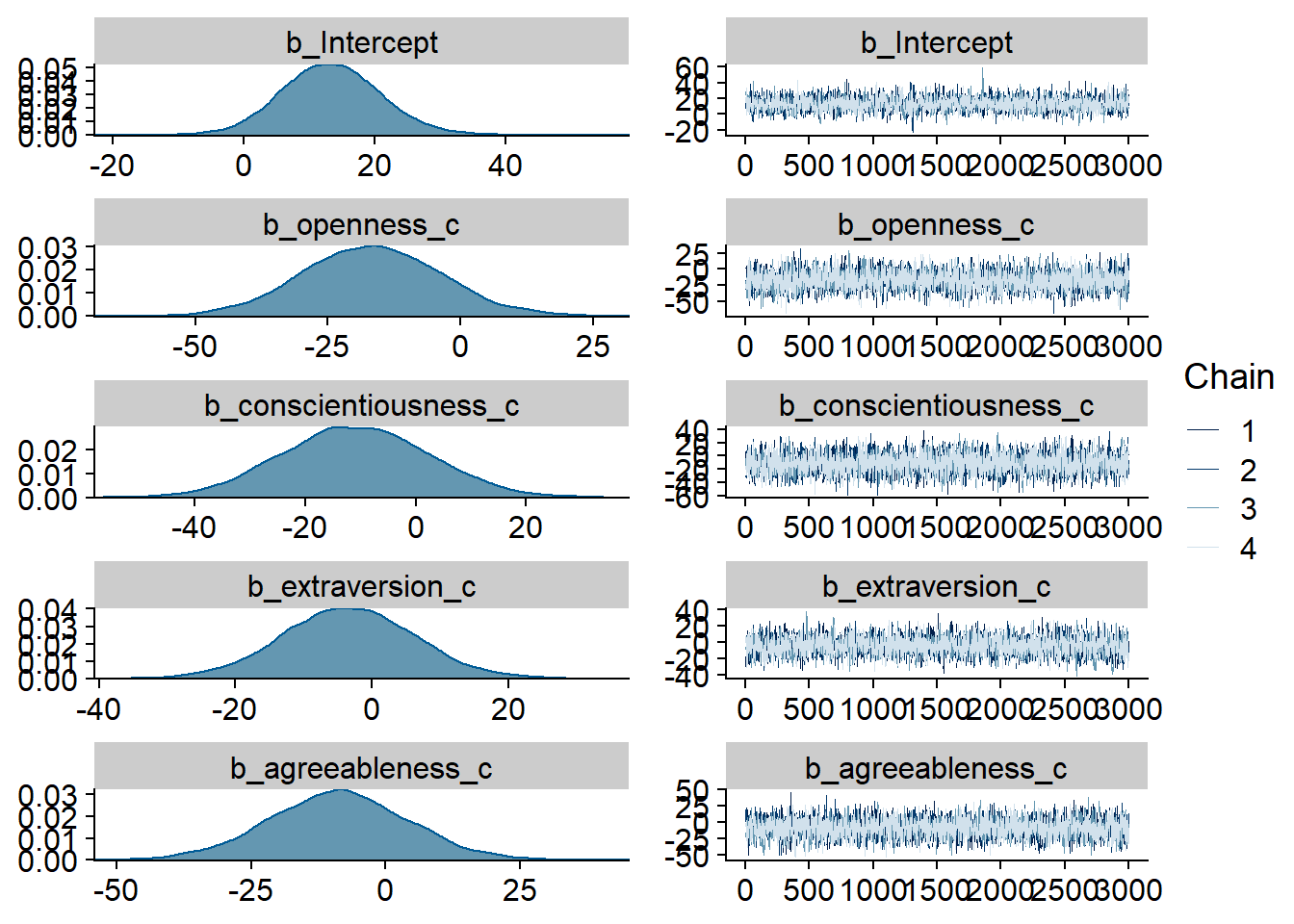
Figure 4.13: Traceplots and posterior distributions for Model 3
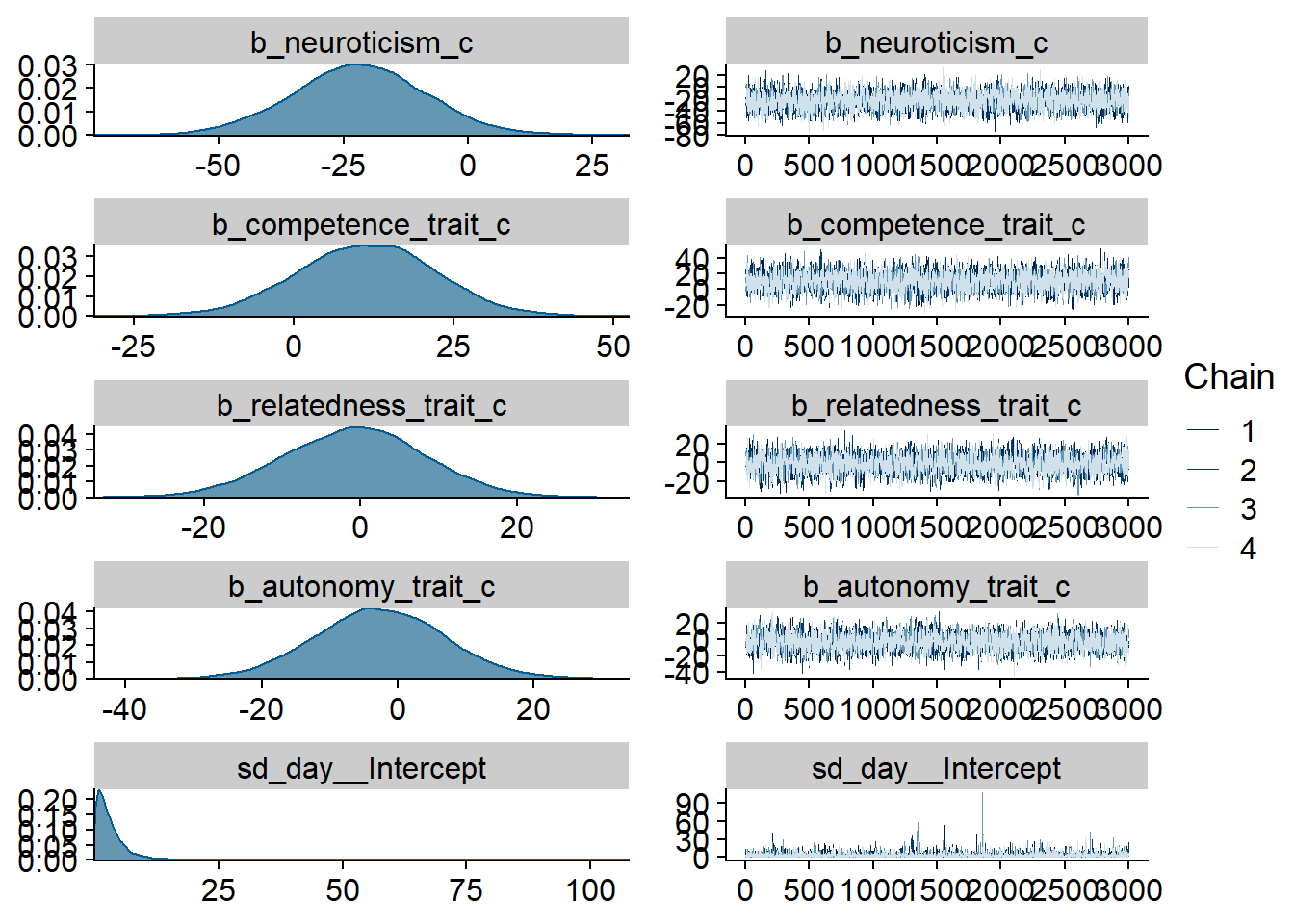
Figure 4.14: Traceplots and posterior distributions for Model 3
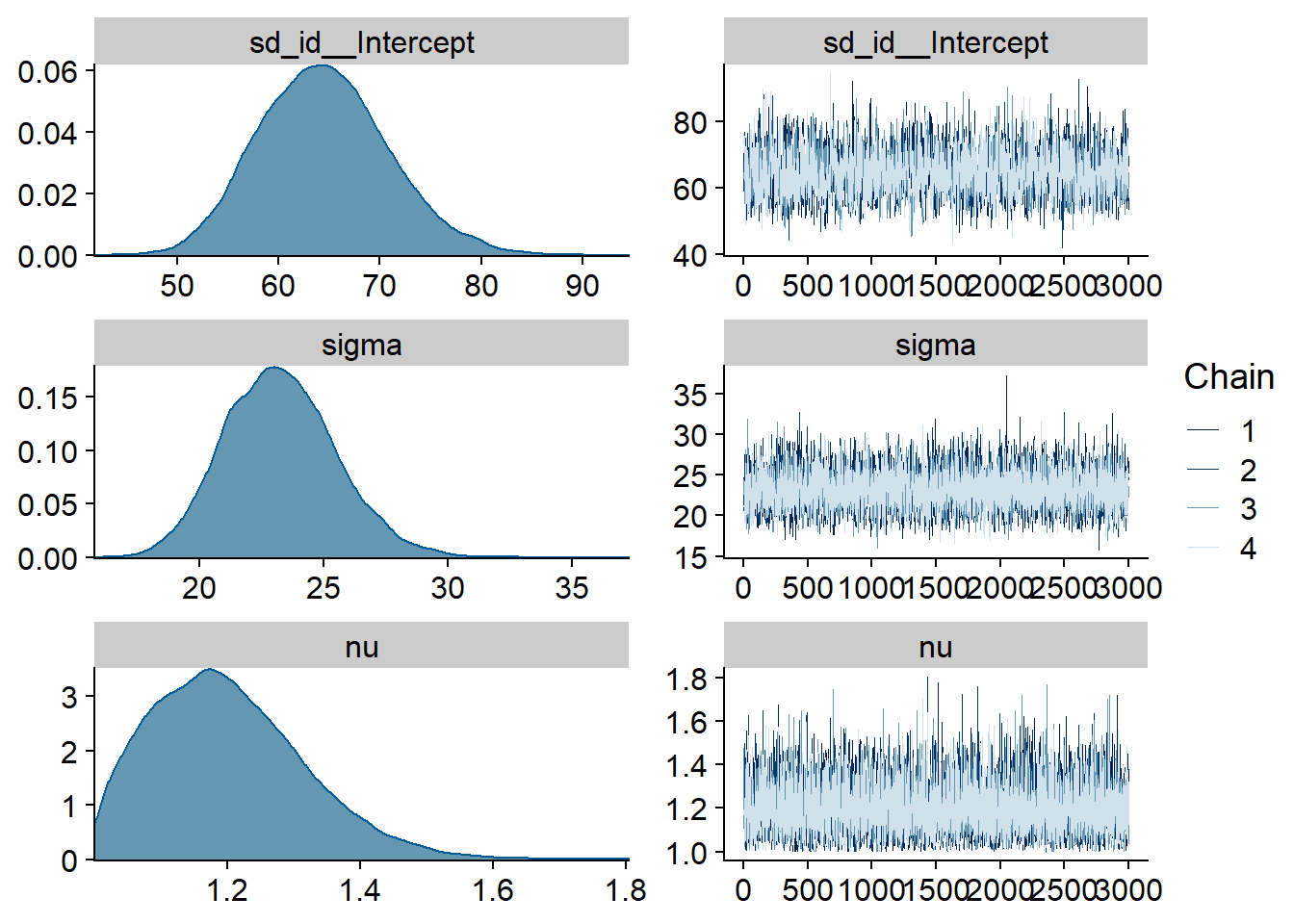
Figure 4.15: Traceplots and posterior distributions for Model 3
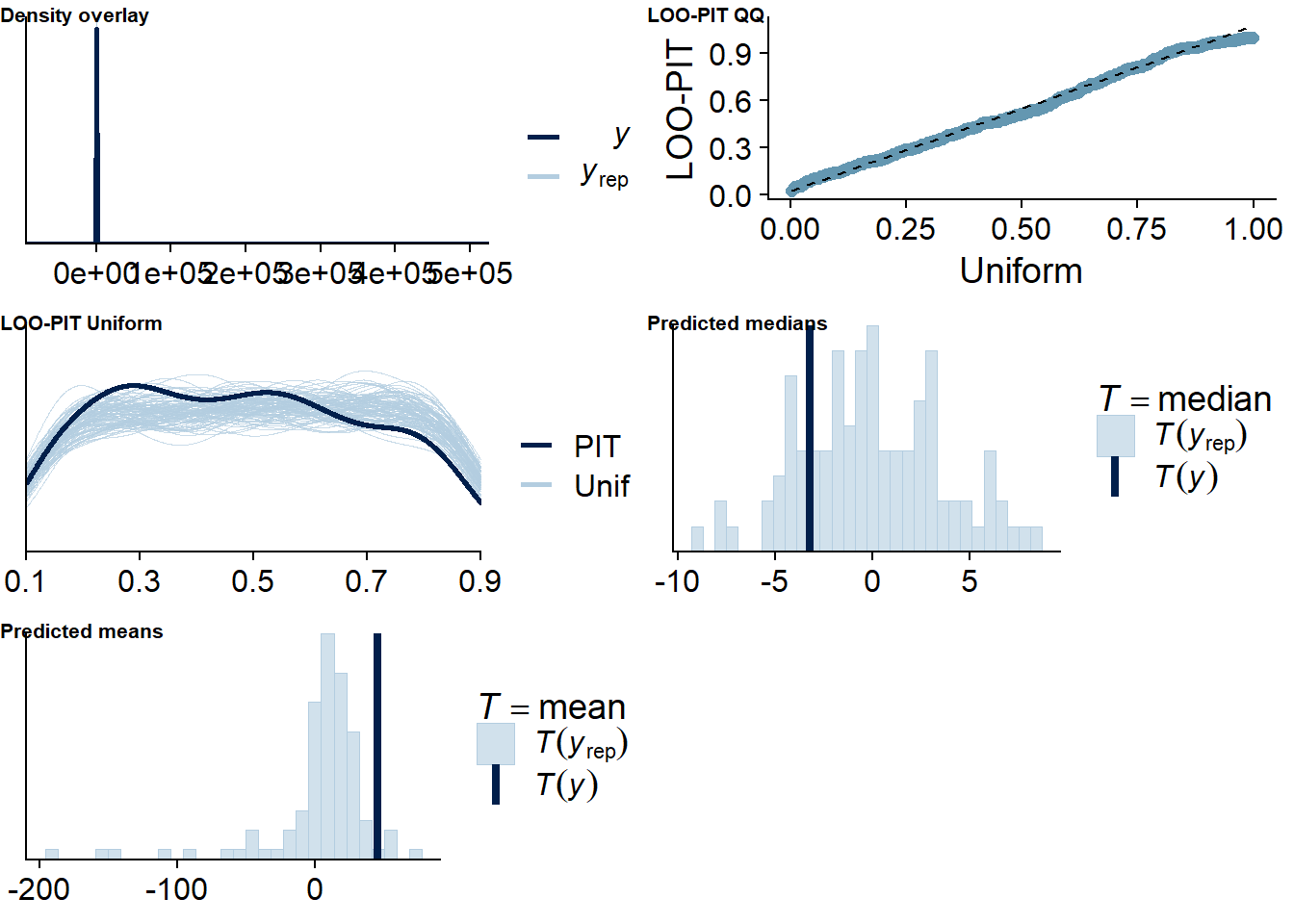
Figure 4.16: Posterior predictive checks for Model 3
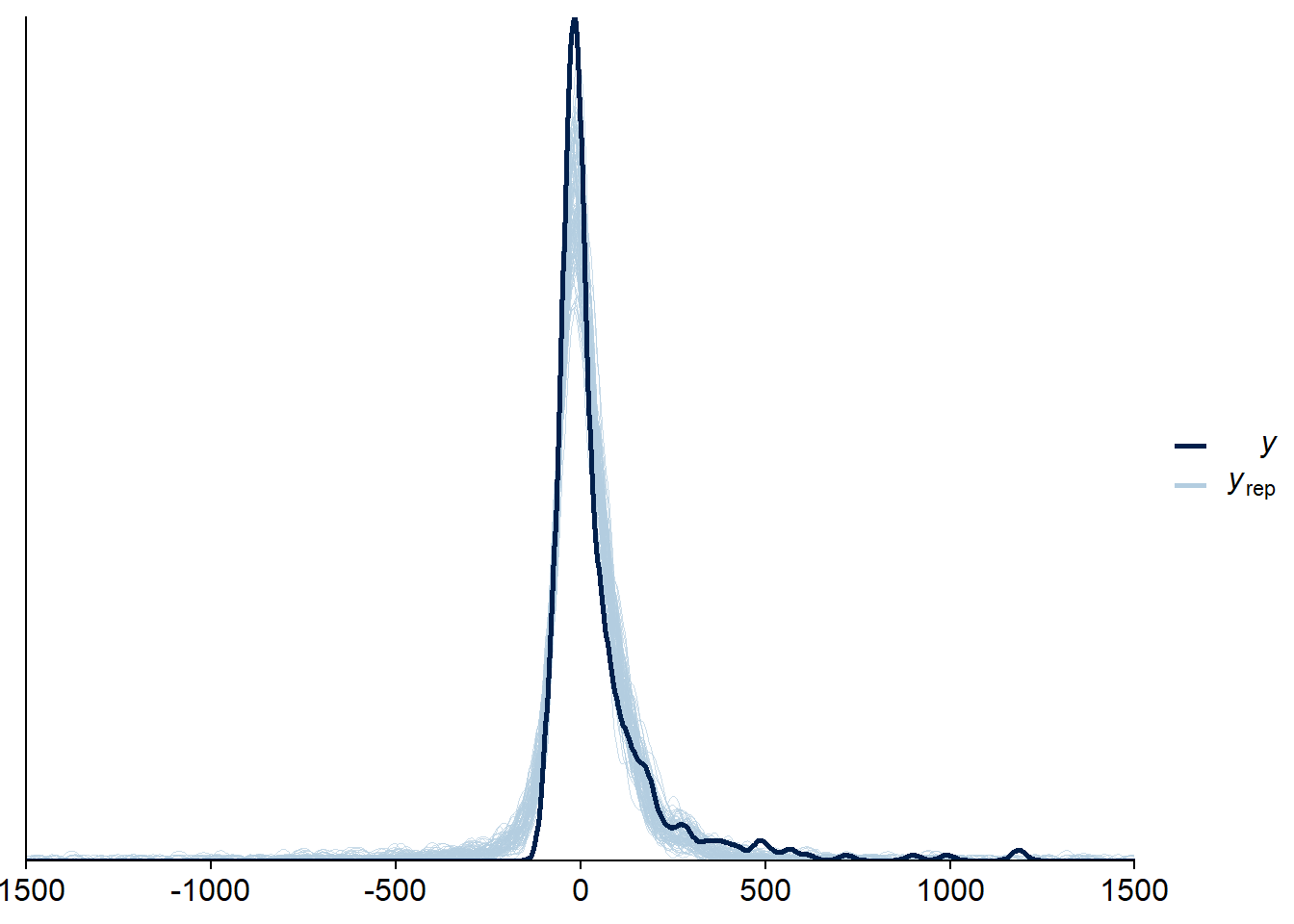
Figure 4.17: Posterior predictive checks for Model 3
Let’s again check for potentially influential values.
Three potentially influential cases are flagged, which is why we calculate them precisely by setting reloo to TRUE.
When we calculate ELPD directly, all values appear unproblematic.
The results seem trustworthy.
## 3 problematic observation(s) found.
## The model will be refit 3 times.##
## Fitting model 1 out of 3 (leaving out observation 40)##
## Fitting model 2 out of 3 (leaving out observation 43)##
## Fitting model 3 out of 3 (leaving out observation 111)## Start sampling
## Start sampling
## Start sampling##
## Computed from 12000 by 426 log-likelihood matrix
##
## Estimate SE
## elpd_loo -2415.2 31.4
## p_loo 136.1 6.7
## looic 4830.3 62.7
## ------
## Monte Carlo SE of elpd_loo is 0.2.
##
## Pareto k diagnostic values:
## Count Pct. Min. n_eff
## (-Inf, 0.5] (good) 421 98.8% 436
## (0.5, 0.7] (ok) 5 1.2% 714
## (0.7, 1] (bad) 0 0.0% <NA>
## (1, Inf) (very bad) 0 0.0% <NA>
##
## All Pareto k estimates are ok (k < 0.7).
## See help('pareto-k-diagnostic') for details.Let’s inspect the summary. There doesn’t seem to be much going on when it comes to the predictors. For all predictors, we cannot be 95% certain that zero isn’t the true effect (conditional on the model). Again, neuroticism comes close.
## Family: student
## Links: mu = identity; sigma = identity; nu = identity
## Formula: error ~ 1 + openness_c + conscientiousness_c + extraversion_c + agreeableness_c + neuroticism_c + competence_trait_c + relatedness_trait_c + autonomy_trait_c + (1 | id) + (1 | day)
## Data: study1 (Number of observations: 426)
## Samples: 4 chains, each with iter = 5000; warmup = 2000; thin = 1;
## total post-warmup samples = 12000
##
## Priors:
## b ~ normal(0, 25)
## Intercept ~ student_t(10, 20, 100)
## nu ~ gamma(2, 0.1)
## sd ~ student_t(3, 0, 57.7)
## sigma ~ student_t(3, 0, 57.7)
##
## Group-Level Effects:
## ~day (Number of levels: 5)
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## sd(Intercept) 3.21 3.59 0.11 11.79 1.00 5317 5339
##
## ~id (Number of levels: 94)
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## sd(Intercept) 64.47 6.40 52.80 77.92 1.00 2251 3791
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## Intercept 13.34 7.62 -1.10 28.66 1.00 2110 4056
## openness_c -16.81 13.17 -42.62 9.80 1.00 2867 4647
## conscientiousness_c -10.92 13.30 -36.68 15.14 1.00 2861 5033
## extraversion_c -3.45 9.78 -22.74 16.05 1.00 2531 4145
## agreeableness_c -8.85 12.86 -34.32 16.69 1.00 2758 4088
## neuroticism_c -21.89 13.39 -48.27 4.58 1.00 2828 4561
## competence_trait_c 11.31 10.95 -10.34 32.57 1.00 2710 4601
## relatedness_trait_c -0.79 9.09 -18.85 16.70 1.00 2056 3941
## autonomy_trait_c -2.66 9.59 -21.25 16.33 1.00 2594 4595
##
## Family Specific Parameters:
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## sigma 23.26 2.23 19.16 27.85 1.00 5374 7711
## nu 1.20 0.11 1.02 1.46 1.00 5581 4487
##
## Samples were drawn using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).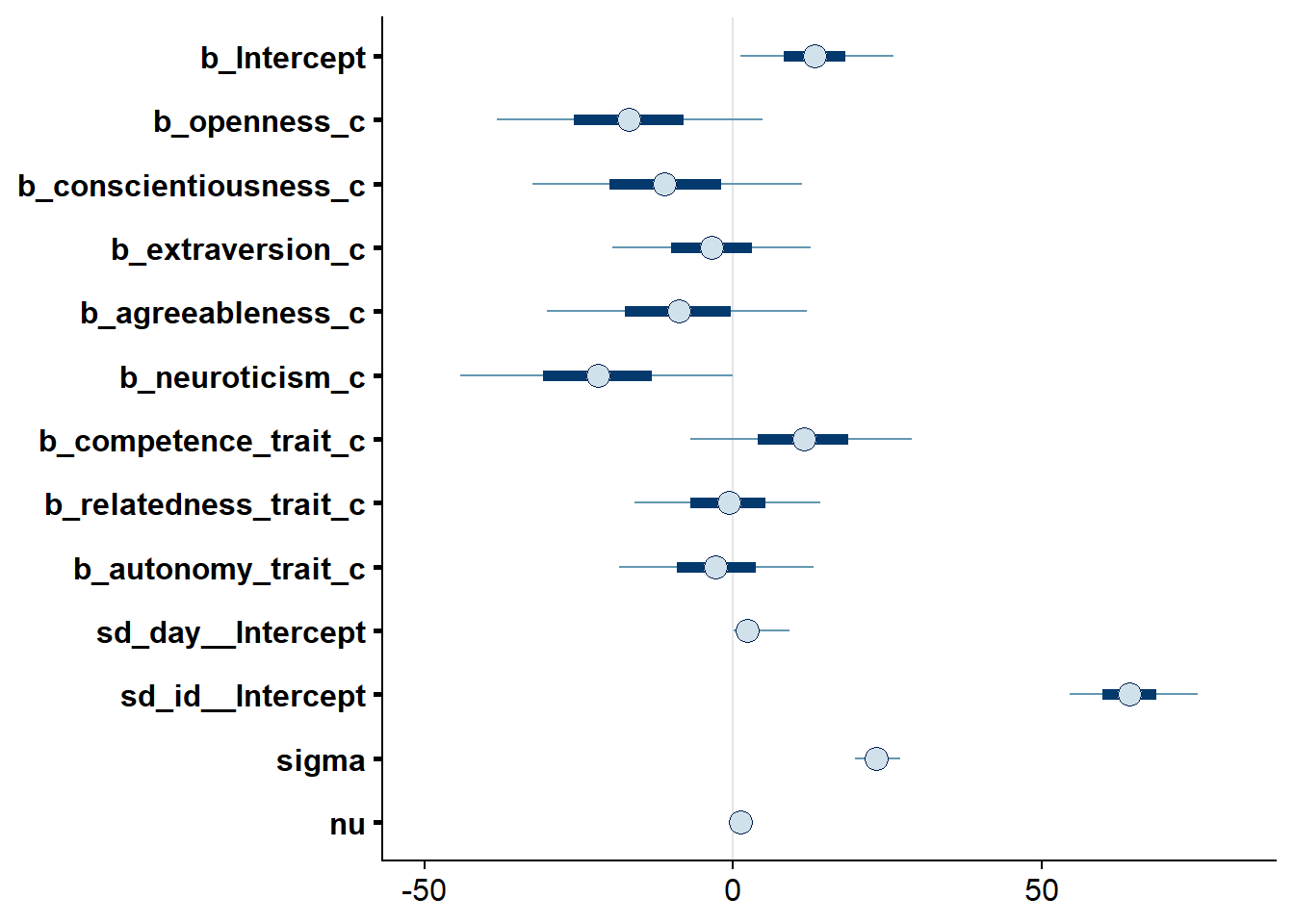
Figure 4.18: Effects plot for Model 3
4.4 Do state (i.e., day-level) variables predict social media use and accuracy?
For the next section, we look at whether day-level variables (i.e., variables reported during experience sampling) predict subjective use, objective use, and accuracy. Again, we have three questions:
- Do day-level variables predict objective-only engagement?
- Do day-level variables predict subjective-only engagement?
- Do day-level variables predict accuracy?
Once more I’ll construct a model for each of those three questions.
Our dependent variables are the same as in the previous block of models: social_media_objective, social_media_subjective, and error.
Our predictors will be variables on the day-level, so state variables: need satisfaction for autonomy, competence, and relatedness as well as four experiential qualities during participants’ days (boredom, enjoyment, satisfaction, and stress).
Because we want to separate between-person and within-person effects, we’ll do group-mean centering (i.e., per participant) and the calculate the deviation of each observation from that group mean. We enter both the person mean and their deviation as predictors, which will lead to a lot of variables, because each of the seven predictors will be separated into a between-person and a within-person predictor. See this blogpost for a tutorial on centering.
dat <-
dat %>%
group_by(id) %>%
mutate(
across(
c(autonomy_state:relatedness_state, satisfied:enjoyable),
list(
between = ~ mean(.x, na.rm = TRUE),
within = ~.x - mean(.x, na.rm = TRUE)
)
)
) %>%
ungroup()4.4.1 Model 4: State variables predicting objective use
First, I choose sensible priors. In contrast to previous models, there isn’t much literature that could inform priors on our predictors. The same goes for the experiential qualities. We’ll go with a “maximal” model where each participant and day get their own intercept plus random slopes nested within participant, because it’s plausible that the effects vary per participant. They could also vary by day, but a) there was little to no variation for day in previous models, and b) that would lead to more parameters than our little data could handle. Just like before we assume a Gamma distribution for the social media variables.
I’ll go step-by-step:
- For the intercepts, I use the same prior as above in Models 1 and 2.
- For basic psychological needs fixed effects, this paper reports very small relations between self-determined motivations at work and social media use (\(\beta\) < .06). However, most of the literature focuses on those motivations and social media addiction, enjoyment of social media, or satisfaction with social media. Therefore, I’ll use weakly regularizing priors for those effects, which means I’ll go with the same prior for the effects as for Models 1 and 2. I would expect larger differences on the between-level, based on the literature on media use and well-being. However, we don’t have that info for our predictors and the priors are rather weak, so I’ll apply them to both between and within predictors.
- For the experiential qualities, there isn’t much literature out there that would allow choosing an informed prior. Most of those experiential qualities are about having a fulfilled and good day, which taps into the whole controversy over the relation between such experiences and social media. Therefore, I’ll take a skeptical stance here and again assign the same weakly regularizing priors (aka the priors we also used for Models 1 and 2).
- For the variances (sigmas) I’ll take the
brmsdefault priors because I have no prior information, nor how the effects vary across day and participant. - For the correlation between intercepts and slopes, I’ll again use the default prior, mostly because those have shown to help with convergence and I don’t have good information on which correlation to expect.
priors_model4 <-
c(
# intercept
prior(normal(4.5, 0.8), class = Intercept),
# prior on effects
prior(normal(0, 0.1), class = b),
# prior on shape
prior(gamma(2.5, 100), class = shape)
)Let’s run the model. Note that the within-deviations can vary per participant, but not the between effects.
model4 <-
brm(
data = dat,
family = Gamma(link = "log"),
prior = priors_model4,
social_media_objective ~
1 +
autonomy_state_between +
competence_state_between +
relatedness_state_between +
satisfied_between +
boring_between +
stressful_between +
enjoyable_between +
autonomy_state_within +
competence_state_within +
relatedness_state_within +
satisfied_within +
boring_within +
stressful_within +
enjoyable_within +
(
1 +
autonomy_state_within +
competence_state_within +
relatedness_state_within +
satisfied_within +
boring_within +
stressful_within +
enjoyable_within |
id
) +
(1 |day),
iter = 5000,
warmup = 2000,
chains = 4,
cores = 4,
seed = 42,
control = list(
adapt_delta = 0.99
),
file = here("models", "model4")
)
Figure 4.19: Traceplots and posterior distributions for Model 4
Figure 4.20: Traceplots and posterior distributions for Model 4
Figure 4.21: Traceplots and posterior distributions for Model 4
Figure 4.22: Traceplots and posterior distributions for Model 4
Figure 4.23: Traceplots and posterior distributions for Model 4
Figure 4.24: Traceplots and posterior distributions for Model 4
Figure 4.25: Traceplots and posterior distributions for Model 4
Figure 4.26: Traceplots and posterior distributions for Model 4
Figure 4.27: Traceplots and posterior distributions for Model 4
Figure 4.28: Traceplots and posterior distributions for Model 4
Figure 4.29: Traceplots and posterior distributions for Model 4
Figure 4.30: Posterior predictive checks for Model 4
Next, we check for potentially influential cases. There are none.
##
## Computed from 12000 by 420 log-likelihood matrix
##
## Estimate SE
## elpd_loo -2412.8 14.1
## p_loo 25.6 1.6
## looic 4825.6 28.1
## ------
## Monte Carlo SE of elpd_loo is 0.1.
##
## Pareto k diagnostic values:
## Count Pct. Min. n_eff
## (-Inf, 0.5] (good) 408 97.1% 4830
## (0.5, 0.7] (ok) 12 2.9% 1078
## (0.7, 1] (bad) 0 0.0% <NA>
## (1, Inf) (very bad) 0 0.0% <NA>
##
## All Pareto k estimates are ok (k < 0.7).
## See help('pareto-k-diagnostic') for details.Let’s inspect the summary. There doesn’t seem to be much going on when it comes to the predictors.
## Family: gamma
## Links: mu = log; shape = identity
## Formula: social_media_objective ~ 1 + autonomy_state_between + competence_state_between + relatedness_state_between + satisfied_between + boring_between + stressful_between + enjoyable_between + autonomy_state_within + competence_state_within + relatedness_state_within + satisfied_within + boring_within + stressful_within + enjoyable_within + (1 + autonomy_state_within + competence_state_within + relatedness_state_within + satisfied_within + boring_within + stressful_within + enjoyable_within | id) + (1 | day)
## Data: study1 (Number of observations: 420)
## Samples: 4 chains, each with iter = 5000; warmup = 2000; thin = 1;
## total post-warmup samples = 12000
##
## Priors:
## b ~ normal(0, 0.1)
## Intercept ~ normal(4.5, 0.8)
## L ~ lkj_corr_cholesky(1)
## sd ~ student_t(3, 0, 2.5)
## shape ~ gamma(2.5, 100)
##
## Group-Level Effects:
## ~day (Number of levels: 5)
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## sd(Intercept) 0.07 0.07 0.00 0.25 1.00 5832 6519
##
## ~id (Number of levels: 94)
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## sd(Intercept) 0.52 0.08 0.38 0.67 1.00 3573 5944
## sd(autonomy_state_within) 0.05 0.04 0.00 0.16 1.00 8320 5372
## sd(competence_state_within) 0.05 0.04 0.00 0.14 1.00 8935 5381
## sd(relatedness_state_within) 0.07 0.05 0.00 0.19 1.00 7787 4916
## sd(satisfied_within) 0.04 0.03 0.00 0.12 1.00 8933 5890
## sd(boring_within) 0.03 0.03 0.00 0.10 1.00 9702 5442
## sd(stressful_within) 0.03 0.02 0.00 0.09 1.00 9082 6157
## sd(enjoyable_within) 0.04 0.03 0.00 0.12 1.00 9115 5934
## cor(Intercept,autonomy_state_within) -0.02 0.33 -0.65 0.61 1.00 16387 8961
## cor(Intercept,competence_state_within) -0.01 0.34 -0.64 0.63 1.00 21044 9001
## cor(autonomy_state_within,competence_state_within) -0.04 0.34 -0.66 0.60 1.00 13676 9802
## cor(Intercept,relatedness_state_within) 0.00 0.33 -0.62 0.63 1.00 20886 8610
## cor(autonomy_state_within,relatedness_state_within) -0.02 0.33 -0.65 0.61 1.00 14368 8242
## cor(competence_state_within,relatedness_state_within) -0.02 0.33 -0.65 0.62 1.00 11400 8640
## cor(Intercept,satisfied_within) -0.02 0.33 -0.65 0.62 1.00 19647 8386
## cor(autonomy_state_within,satisfied_within) -0.03 0.33 -0.65 0.62 1.00 13873 8112
## cor(competence_state_within,satisfied_within) -0.03 0.34 -0.66 0.60 1.00 11687 9023
## cor(relatedness_state_within,satisfied_within) -0.03 0.33 -0.65 0.62 1.00 10544 9670
## cor(Intercept,boring_within) 0.05 0.33 -0.60 0.67 1.00 21420 8481
## cor(autonomy_state_within,boring_within) 0.02 0.34 -0.62 0.64 1.00 16012 9390
## cor(competence_state_within,boring_within) 0.02 0.33 -0.61 0.64 1.00 12124 9240
## cor(relatedness_state_within,boring_within) 0.01 0.33 -0.62 0.63 1.00 9880 9056
## cor(satisfied_within,boring_within) 0.01 0.34 -0.63 0.64 1.00 9165 9884
## cor(Intercept,stressful_within) 0.01 0.33 -0.63 0.64 1.00 18727 8771
## cor(autonomy_state_within,stressful_within) 0.04 0.34 -0.61 0.66 1.00 14611 8703
## cor(competence_state_within,stressful_within) 0.03 0.33 -0.62 0.65 1.00 11626 9342
## cor(relatedness_state_within,stressful_within) 0.01 0.33 -0.62 0.65 1.00 10591 9391
## cor(satisfied_within,stressful_within) 0.01 0.33 -0.62 0.64 1.00 8292 9818
## cor(boring_within,stressful_within) -0.02 0.33 -0.64 0.62 1.00 8189 9968
## cor(Intercept,enjoyable_within) -0.04 0.33 -0.66 0.62 1.00 19274 7944
## cor(autonomy_state_within,enjoyable_within) -0.04 0.34 -0.68 0.62 1.00 13860 8987
## cor(competence_state_within,enjoyable_within) -0.03 0.34 -0.66 0.61 1.00 11180 8545
## cor(relatedness_state_within,enjoyable_within) -0.02 0.34 -0.65 0.63 1.00 9846 9047
## cor(satisfied_within,enjoyable_within) -0.05 0.33 -0.66 0.61 1.00 8402 9749
## cor(boring_within,enjoyable_within) 0.03 0.33 -0.61 0.65 1.00 7006 9546
## cor(stressful_within,enjoyable_within) 0.02 0.33 -0.63 0.64 1.00 7054 9355
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## Intercept 4.61 0.66 3.32 5.89 1.00 7471 8345
## autonomy_state_between -0.02 0.08 -0.17 0.12 1.00 9979 9622
## competence_state_between 0.01 0.07 -0.13 0.15 1.00 9383 10215
## relatedness_state_between -0.01 0.07 -0.15 0.14 1.00 9084 9159
## satisfied_between -0.04 0.07 -0.18 0.11 1.00 10538 9759
## boring_between 0.05 0.06 -0.07 0.16 1.00 7557 9219
## stressful_between 0.01 0.05 -0.10 0.11 1.00 7654 9029
## enjoyable_between 0.07 0.07 -0.07 0.22 1.00 9402 8694
## autonomy_state_within -0.01 0.06 -0.12 0.10 1.00 16434 8971
## competence_state_within -0.06 0.06 -0.17 0.05 1.00 15497 9011
## relatedness_state_within 0.01 0.06 -0.11 0.13 1.00 19434 9421
## satisfied_within 0.02 0.05 -0.08 0.11 1.00 15316 8718
## boring_within -0.01 0.04 -0.09 0.06 1.00 15776 9182
## stressful_within -0.02 0.04 -0.09 0.05 1.00 16505 9577
## enjoyable_within 0.03 0.05 -0.06 0.12 1.00 14243 9107
##
## Family Specific Parameters:
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## shape 1.41 0.10 1.22 1.62 1.00 8991 8328
##
## Samples were drawn using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).
Figure 4.31: Effects plot for Model 4
4.4.2 Model 5: State variables predicting subjective use
The next model predicts subjective use from the same predictors. Once more, we know that subjective and objective use aren’t perfectly correlated, but the priors for Model 4 again are our best bet here (for both within-person and between-person effects).
priors_model5 <-
c(
# intercept
prior(normal(4.5, 0.8), class = Intercept),
# prior on effects
prior(normal(0, 0.1), class = b),
# prior on shape
prior(gamma(2.5, 100), class = shape)
)Let’s run the model. Like with Model 2, we don’t separately model zeros.
model5 <-
brm(
data = dat,
family = Gamma(link = "log"),
prior = priors_model4,
social_media_subjective ~
1 +
autonomy_state_between +
competence_state_between +
relatedness_state_between +
satisfied_between +
boring_between +
stressful_between +
enjoyable_between +
autonomy_state_within +
competence_state_within +
relatedness_state_within +
satisfied_within +
boring_within +
stressful_within +
enjoyable_within +
(
1 +
autonomy_state_within +
competence_state_within +
relatedness_state_within +
satisfied_within +
boring_within +
stressful_within +
enjoyable_within |
id
) +
(1 |day),
iter = 5000,
warmup = 2000,
chains = 4,
cores = 4,
seed = 42,
control = list(
adapt_delta = 0.99
),
file = here("models", "model5")
)
Figure 4.32: Traceplots and posterior distributions for Model 4
Figure 4.33: Traceplots and posterior distributions for Model 4
Figure 4.34: Traceplots and posterior distributions for Model 4
Figure 4.35: Traceplots and posterior distributions for Model 4
Figure 4.36: Traceplots and posterior distributions for Model 4
Figure 4.37: Traceplots and posterior distributions for Model 4
Figure 4.38: Traceplots and posterior distributions for Model 4
Figure 4.39: Traceplots and posterior distributions for Model 4
Figure 4.40: Traceplots and posterior distributions for Model 4
Figure 4.41: Traceplots and posterior distributions for Model 4
Figure 4.42: Traceplots and posterior distributions for Model 4
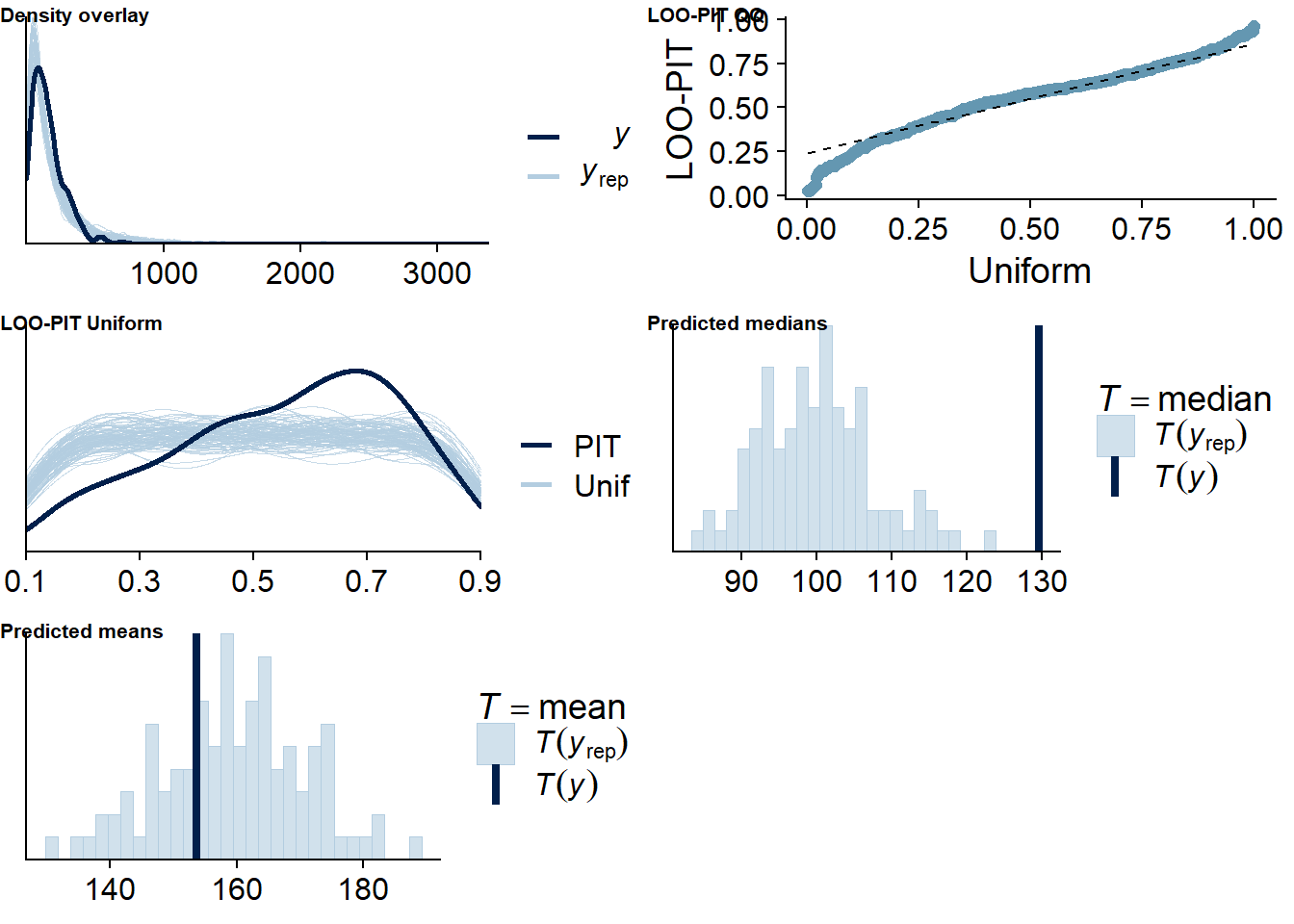
Figure 4.43: Posterior predictive checks for Model 5
There are no cases flagged as potentially influential outliers.
## 1 problematic observation(s) found.
## The model will be refit 1 times.##
## Fitting model 1 out of 1 (leaving out observation 384)## Start sampling##
## Computed from 12000 by 420 log-likelihood matrix
##
## Estimate SE
## elpd_loo -2466.6 15.8
## p_loo 28.8 1.6
## looic 4933.2 31.6
## ------
## Monte Carlo SE of elpd_loo is 0.1.
##
## Pareto k diagnostic values:
## Count Pct. Min. n_eff
## (-Inf, 0.5] (good) 393 93.6% 1996
## (0.5, 0.7] (ok) 27 6.4% 1886
## (0.7, 1] (bad) 0 0.0% <NA>
## (1, Inf) (very bad) 0 0.0% <NA>
##
## All Pareto k estimates are ok (k < 0.7).
## See help('pareto-k-diagnostic') for details.Let’s inspect the summary. Having an enjoyable and boring day might be related to estimating more social media use (on the between level). Conversely, a feeling of competence might decrease estimates. However, all three of those posterior distributions include zero, even though it’s close.
## Family: gamma
## Links: mu = log; shape = identity
## Formula: social_media_subjective ~ 1 + autonomy_state_between + competence_state_between + relatedness_state_between + satisfied_between + boring_between + stressful_between + enjoyable_between + autonomy_state_within + competence_state_within + relatedness_state_within + satisfied_within + boring_within + stressful_within + enjoyable_within + (1 + autonomy_state_within + competence_state_within + relatedness_state_within + satisfied_within + boring_within + stressful_within + enjoyable_within | id) + (1 | day)
## Data: study1 (Number of observations: 420)
## Samples: 4 chains, each with iter = 5000; warmup = 2000; thin = 1;
## total post-warmup samples = 12000
##
## Priors:
## b ~ normal(0, 0.1)
## Intercept ~ normal(4.5, 0.8)
## L ~ lkj_corr_cholesky(1)
## sd ~ student_t(3, 0, 2.5)
## shape ~ gamma(2.5, 100)
##
## Group-Level Effects:
## ~day (Number of levels: 5)
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## sd(Intercept) 0.07 0.08 0.00 0.26 1.00 6115 6209
##
## ~id (Number of levels: 94)
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## sd(Intercept) 0.49 0.08 0.35 0.64 1.00 4609 6905
## sd(autonomy_state_within) 0.06 0.04 0.00 0.16 1.00 9124 6247
## sd(competence_state_within) 0.05 0.04 0.00 0.16 1.00 9417 5712
## sd(relatedness_state_within) 0.07 0.05 0.00 0.20 1.00 10131 6334
## sd(satisfied_within) 0.05 0.04 0.00 0.14 1.00 8222 5408
## sd(boring_within) 0.04 0.03 0.00 0.11 1.00 9518 5873
## sd(stressful_within) 0.04 0.03 0.00 0.10 1.00 9568 6523
## sd(enjoyable_within) 0.04 0.03 0.00 0.13 1.00 8525 6408
## cor(Intercept,autonomy_state_within) -0.02 0.34 -0.65 0.63 1.00 26697 8853
## cor(Intercept,competence_state_within) -0.03 0.33 -0.65 0.61 1.00 25740 8665
## cor(autonomy_state_within,competence_state_within) -0.04 0.34 -0.65 0.61 1.00 17150 9376
## cor(Intercept,relatedness_state_within) 0.01 0.33 -0.63 0.65 1.00 27531 8584
## cor(autonomy_state_within,relatedness_state_within) -0.02 0.34 -0.66 0.62 1.00 16808 9191
## cor(competence_state_within,relatedness_state_within) -0.02 0.33 -0.64 0.61 1.00 12371 9241
## cor(Intercept,satisfied_within) 0.01 0.33 -0.62 0.64 1.00 23477 8466
## cor(autonomy_state_within,satisfied_within) -0.03 0.33 -0.66 0.62 1.00 16534 8899
## cor(competence_state_within,satisfied_within) -0.03 0.34 -0.67 0.62 1.00 12832 9212
## cor(relatedness_state_within,satisfied_within) -0.03 0.33 -0.65 0.61 1.00 10261 10017
## cor(Intercept,boring_within) 0.04 0.33 -0.60 0.65 1.00 21811 8984
## cor(autonomy_state_within,boring_within) 0.03 0.33 -0.61 0.66 1.00 17402 9064
## cor(competence_state_within,boring_within) 0.02 0.33 -0.62 0.65 1.00 13863 8855
## cor(relatedness_state_within,boring_within) 0.01 0.34 -0.63 0.64 1.00 10166 9533
## cor(satisfied_within,boring_within) 0.02 0.33 -0.61 0.64 1.00 9338 9412
## cor(Intercept,stressful_within) 0.04 0.33 -0.59 0.64 1.00 25952 7970
## cor(autonomy_state_within,stressful_within) 0.03 0.33 -0.62 0.66 1.00 15050 8199
## cor(competence_state_within,stressful_within) 0.03 0.34 -0.62 0.66 1.00 12065 8725
## cor(relatedness_state_within,stressful_within) 0.01 0.33 -0.62 0.64 1.00 9962 9381
## cor(satisfied_within,stressful_within) 0.01 0.33 -0.62 0.63 1.00 9639 9836
## cor(boring_within,stressful_within) -0.01 0.33 -0.64 0.62 1.00 7948 9156
## cor(Intercept,enjoyable_within) -0.05 0.33 -0.65 0.59 1.00 24227 9637
## cor(autonomy_state_within,enjoyable_within) -0.03 0.34 -0.66 0.61 1.00 15460 8292
## cor(competence_state_within,enjoyable_within) -0.03 0.33 -0.65 0.60 1.00 12580 8950
## cor(relatedness_state_within,enjoyable_within) -0.02 0.33 -0.64 0.61 1.00 11135 9699
## cor(satisfied_within,enjoyable_within) -0.04 0.34 -0.67 0.60 1.00 8582 9049
## cor(boring_within,enjoyable_within) 0.03 0.33 -0.62 0.65 1.00 8034 9232
## cor(stressful_within,enjoyable_within) 0.02 0.34 -0.63 0.66 1.00 6775 10061
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## Intercept 5.36 0.66 4.06 6.63 1.00 11737 10046
## autonomy_state_between -0.07 0.08 -0.22 0.08 1.00 13941 8408
## competence_state_between -0.09 0.07 -0.23 0.05 1.00 12371 9819
## relatedness_state_between 0.01 0.07 -0.14 0.15 1.00 14157 8970
## satisfied_between -0.10 0.08 -0.25 0.05 1.00 13708 8537
## boring_between 0.09 0.06 -0.02 0.21 1.00 11432 9251
## stressful_between -0.02 0.05 -0.12 0.08 1.00 10961 9963
## enjoyable_between 0.12 0.08 -0.03 0.26 1.00 13528 9775
## autonomy_state_within -0.00 0.06 -0.12 0.11 1.00 23098 9607
## competence_state_within -0.04 0.06 -0.15 0.07 1.00 19223 8865
## relatedness_state_within -0.00 0.06 -0.13 0.12 1.00 23114 9482
## satisfied_within -0.03 0.05 -0.12 0.07 1.00 18050 9322
## boring_within 0.02 0.04 -0.06 0.10 1.00 21043 9152
## stressful_within -0.02 0.04 -0.10 0.05 1.00 22686 9812
## enjoyable_within 0.04 0.05 -0.06 0.14 1.00 19642 8730
##
## Family Specific Parameters:
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## shape 1.30 0.09 1.12 1.49 1.00 11230 9042
##
## Samples were drawn using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).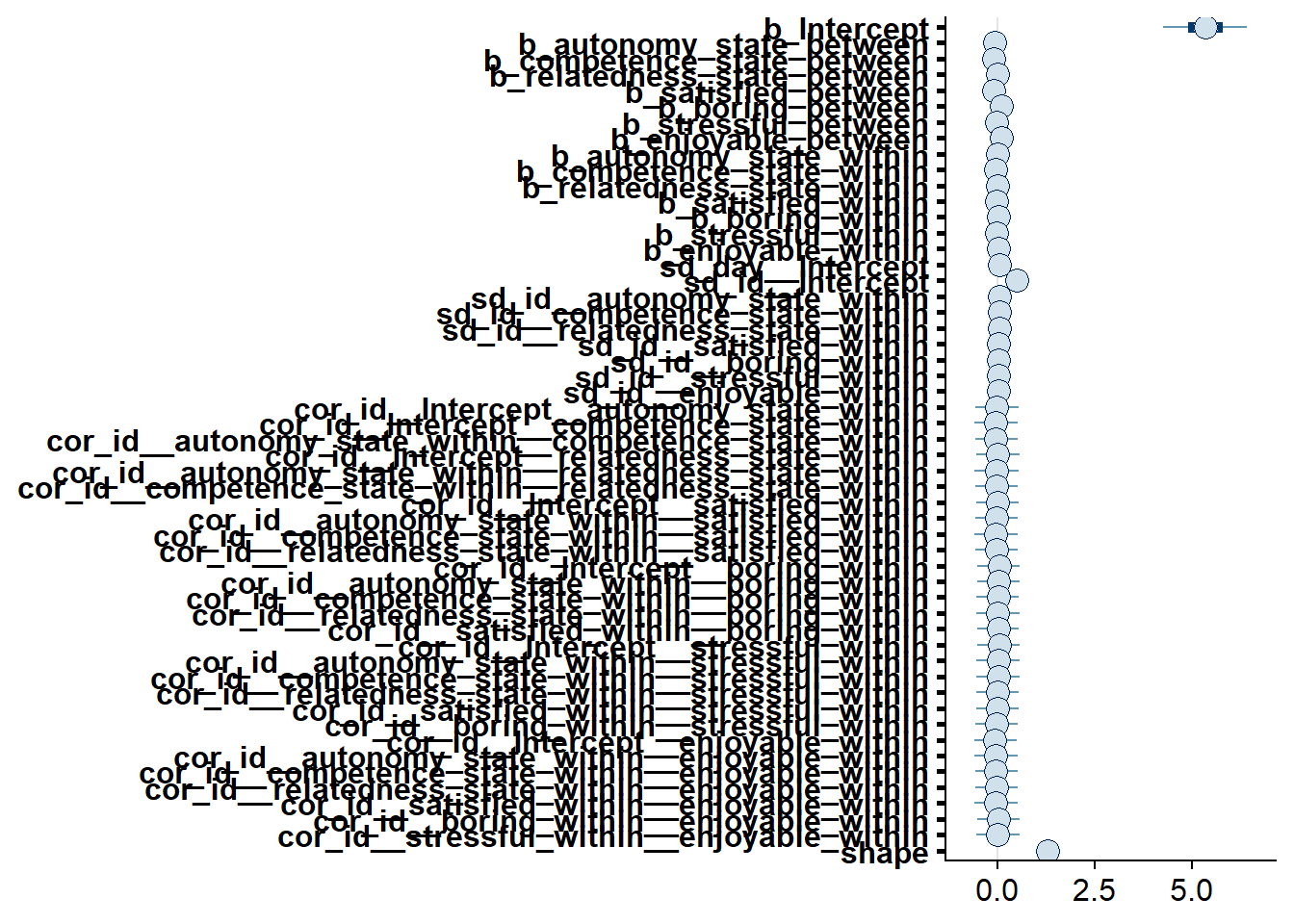
Figure 4.44: Effects plot for Model 5
4.4.3 Model 6: State variables predicting accuracy
For this model, once more it’s hard to choose informed priors.
Therefore, I choose the same skeptical, but weakly regularizing priors as for Model 3, with a student t distribution with fat tails, an intercept that reflects a tendency to overestimate, and slopes that are skeptical of large effects.
For everything else, I’ll use the brms default priors because I don’t have information on what correlation or variances to expect.
Note that I use the priors on the slope for both between and within effects, simply because I don’t have an informed guess about the two being different.
priors_model6 <-
c(
# intercept
prior(student_t(10, 20, 100), class = Intercept),
# all other effects
prior(normal(0, 25), class = b)
)Let’s run the model.
model6 <-
brm(
data = dat,
family = student,
prior = priors_model6,
error ~
1 +
autonomy_state_between +
competence_state_between +
relatedness_state_between +
satisfied_between +
boring_between +
stressful_between +
enjoyable_between +
autonomy_state_within +
competence_state_within +
relatedness_state_within +
satisfied_within +
boring_within +
stressful_within +
enjoyable_within +
(
1 +
autonomy_state_within +
competence_state_within +
relatedness_state_within +
satisfied_within +
boring_within +
stressful_within +
enjoyable_within |
id
) +
(1 |day),
iter = 5000,
warmup = 2000,
chains = 4,
cores = 4,
seed = 42,
control = list(
adapt_delta = 0.99
),
file = here("models", "model6")
)
Figure 4.45: Traceplots and posterior distributions for Model 6
Figure 4.46: Traceplots and posterior distributions for Model 6
Figure 4.47: Traceplots and posterior distributions for Model 6
Figure 4.48: Traceplots and posterior distributions for Model 6
Figure 4.49: Traceplots and posterior distributions for Model 6
Figure 4.50: Traceplots and posterior distributions for Model 6
Figure 4.51: Traceplots and posterior distributions for Model 6
Figure 4.52: Traceplots and posterior distributions for Model 6
Figure 4.53: Traceplots and posterior distributions for Model 6
Figure 4.54: Traceplots and posterior distributions for Model 6
Figure 4.55: Traceplots and posterior distributions for Model 6
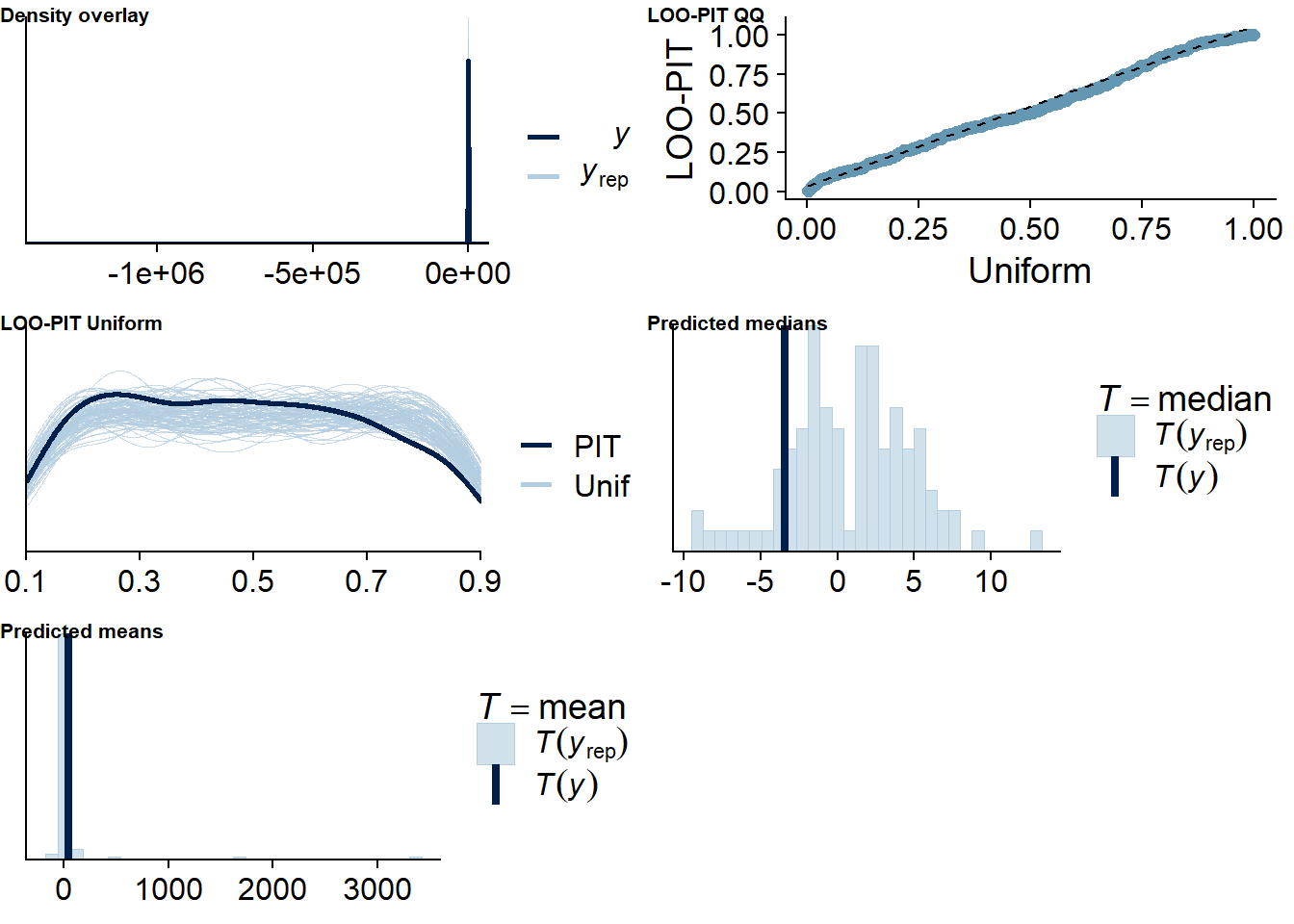
Figure 4.56: Posterior predictive checks for Model 6
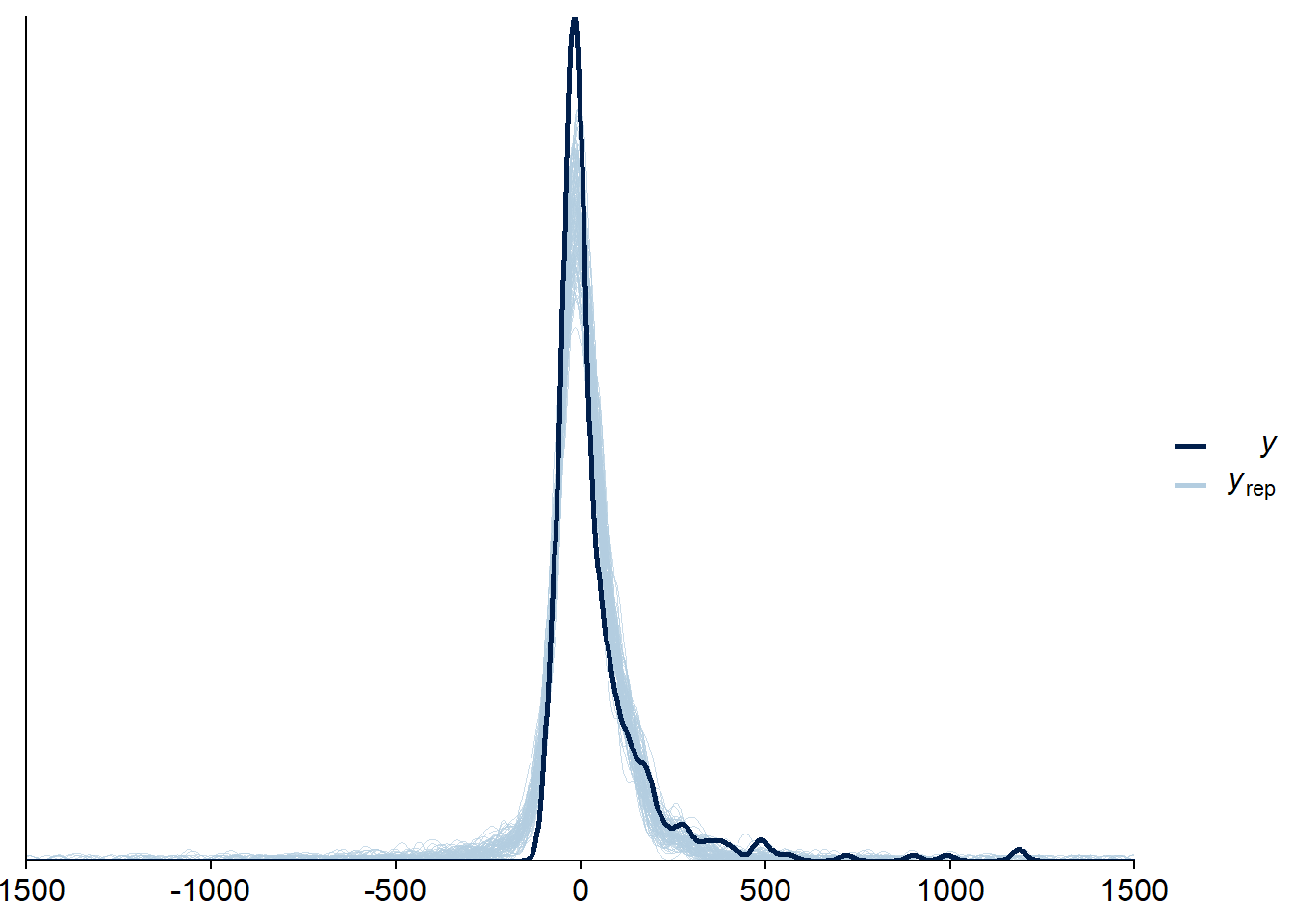
Figure 4.57: Posterior predictive checks for Model 6
Two cases are flagged as potential outliers, which disappear once calculated directly.
## 2 problematic observation(s) found.
## The model will be refit 2 times.##
## Fitting model 1 out of 2 (leaving out observation 39)##
## Fitting model 2 out of 2 (leaving out observation 128)## Start sampling
## Start sampling##
## Computed from 12000 by 418 log-likelihood matrix
##
## Estimate SE
## elpd_loo -2368.1 30.7
## p_loo 204.2 7.8
## looic 4736.3 61.4
## ------
## Monte Carlo SE of elpd_loo is 0.2.
##
## Pareto k diagnostic values:
## Count Pct. Min. n_eff
## (-Inf, 0.5] (good) 415 99.3% 524
## (0.5, 0.7] (ok) 3 0.7% 965
## (0.7, 1] (bad) 0 0.0% <NA>
## (1, Inf) (very bad) 0 0.0% <NA>
##
## All Pareto k estimates are ok (k < 0.7).
## See help('pareto-k-diagnostic') for details.Let’s inspect the summary. All posteriors include 0, only satisfaction (within) comes close.
## Family: student
## Links: mu = identity; sigma = identity; nu = identity
## Formula: error ~ 1 + autonomy_state_between + competence_state_between + relatedness_state_between + satisfied_between + boring_between + stressful_between + enjoyable_between + autonomy_state_within + competence_state_within + relatedness_state_within + satisfied_within + boring_within + stressful_within + enjoyable_within + (1 + autonomy_state_within + competence_state_within + relatedness_state_within + satisfied_within + boring_within + stressful_within + enjoyable_within | id) + (1 | day)
## Data: study1 (Number of observations: 418)
## Samples: 4 chains, each with iter = 5000; warmup = 2000; thin = 1;
## total post-warmup samples = 12000
##
## Priors:
## b ~ normal(0, 25)
## Intercept ~ student_t(10, 20, 100)
## L ~ lkj_corr_cholesky(1)
## nu ~ gamma(2, 0.1)
## sd ~ student_t(3, 0, 56.4)
## sigma ~ student_t(3, 0, 56.4)
##
## Group-Level Effects:
## ~day (Number of levels: 5)
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## sd(Intercept) 3.42 3.53 0.12 12.17 1.00 7456 7118
##
## ~id (Number of levels: 94)
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## sd(Intercept) 61.71 6.13 50.50 74.40 1.00 2533 5226
## sd(autonomy_state_within) 6.08 3.99 0.35 14.85 1.00 3444 5438
## sd(competence_state_within) 3.93 2.98 0.16 11.07 1.00 5720 6710
## sd(relatedness_state_within) 9.65 6.50 0.45 23.82 1.00 1877 3608
## sd(satisfied_within) 7.20 4.24 0.43 16.23 1.00 2343 4739
## sd(boring_within) 2.93 2.28 0.10 8.42 1.00 4427 5807
## sd(stressful_within) 4.68 3.13 0.21 11.56 1.00 1907 4368
## sd(enjoyable_within) 3.54 2.65 0.15 9.98 1.00 4206 6088
## cor(Intercept,autonomy_state_within) 0.03 0.31 -0.57 0.61 1.00 11743 8439
## cor(Intercept,competence_state_within) 0.00 0.33 -0.63 0.64 1.00 15909 8803
## cor(autonomy_state_within,competence_state_within) -0.05 0.34 -0.68 0.60 1.00 12817 9811
## cor(Intercept,relatedness_state_within) -0.14 0.31 -0.68 0.51 1.00 7624 8454
## cor(autonomy_state_within,relatedness_state_within) -0.05 0.33 -0.66 0.60 1.00 7309 8183
## cor(competence_state_within,relatedness_state_within) 0.00 0.33 -0.62 0.63 1.00 7938 9181
## cor(Intercept,satisfied_within) -0.20 0.30 -0.71 0.44 1.00 9855 8185
## cor(autonomy_state_within,satisfied_within) -0.05 0.33 -0.67 0.61 1.00 6581 8107
## cor(competence_state_within,satisfied_within) -0.05 0.33 -0.66 0.61 1.00 6539 8703
## cor(relatedness_state_within,satisfied_within) -0.03 0.33 -0.64 0.61 1.00 7108 8023
## cor(Intercept,boring_within) -0.01 0.33 -0.62 0.62 1.00 15554 9548
## cor(autonomy_state_within,boring_within) -0.01 0.33 -0.64 0.62 1.00 12145 9704
## cor(competence_state_within,boring_within) 0.02 0.33 -0.61 0.64 1.00 10078 9187
## cor(relatedness_state_within,boring_within) -0.00 0.33 -0.63 0.63 1.00 11403 10241
## cor(satisfied_within,boring_within) 0.01 0.33 -0.63 0.63 1.00 11135 10441
## cor(Intercept,stressful_within) 0.03 0.30 -0.56 0.60 1.00 12347 8296
## cor(autonomy_state_within,stressful_within) 0.08 0.34 -0.58 0.69 1.00 6490 7890
## cor(competence_state_within,stressful_within) 0.01 0.33 -0.63 0.64 1.00 6964 8863
## cor(relatedness_state_within,stressful_within) 0.00 0.33 -0.62 0.62 1.00 7347 9079
## cor(satisfied_within,stressful_within) -0.02 0.33 -0.63 0.60 1.00 6808 9360
## cor(boring_within,stressful_within) -0.01 0.33 -0.62 0.62 1.00 7663 9685
## cor(Intercept,enjoyable_within) -0.04 0.32 -0.63 0.59 1.00 16828 8556
## cor(autonomy_state_within,enjoyable_within) -0.04 0.33 -0.66 0.61 1.00 12316 9356
## cor(competence_state_within,enjoyable_within) -0.05 0.34 -0.69 0.61 1.00 10687 9355
## cor(relatedness_state_within,enjoyable_within) -0.04 0.33 -0.66 0.60 1.00 9867 9262
## cor(satisfied_within,enjoyable_within) -0.06 0.33 -0.67 0.60 1.00 10953 10778
## cor(boring_within,enjoyable_within) 0.05 0.34 -0.60 0.68 1.00 9391 10550
## cor(stressful_within,enjoyable_within) 0.05 0.33 -0.60 0.66 1.00 8960 9976
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## Intercept 86.88 77.56 -65.70 238.81 1.00 2752 4530
## autonomy_state_between -3.04 11.00 -24.81 18.66 1.00 3174 5822
## competence_state_between -9.97 9.92 -29.55 9.69 1.00 3299 5130
## relatedness_state_between 5.55 9.55 -13.26 24.35 1.00 3147 5114
## satisfied_between -13.00 11.32 -35.48 9.26 1.00 2544 4848
## boring_between 4.97 6.81 -8.76 18.30 1.00 2619 4431
## stressful_between -7.55 5.83 -18.90 3.92 1.00 3106 5140
## enjoyable_between 6.88 11.12 -14.52 28.89 1.00 2708 5134
## autonomy_state_within -0.25 3.34 -6.83 6.22 1.00 11063 9397
## competence_state_within -0.35 3.15 -6.57 5.92 1.00 10688 8919
## relatedness_state_within 0.48 4.28 -8.21 8.64 1.00 7366 8089
## satisfied_within -6.06 3.09 -12.11 0.06 1.00 9937 9359
## boring_within 2.70 2.01 -1.17 6.75 1.00 9306 9585
## stressful_within -0.49 1.95 -4.34 3.40 1.00 10024 9298
## enjoyable_within 2.63 2.57 -2.37 7.69 1.00 9981 9549
##
## Family Specific Parameters:
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## sigma 20.25 2.41 15.96 25.44 1.00 3054 5285
## nu 1.16 0.11 1.01 1.42 1.00 6060 6060
##
## Samples were drawn using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).
Figure 4.58: Effects plot for Model 6
4.5 Does social media use predict well-being?
For this RQ, we have three questions blocks. We want to know how day-level social media use (both types) and accuracy are related to well-being.
The three questions are: 1. Does objective-only engagement predict day-level well-being? 2. Does subjective-only engagement predict day-level well-being? 3. Does accuracy predict day-level well-being?
4.5.1 Model 7: Objective use predicting well-being
For this model, we’ll predict well-being on that day with objective social media use on that day. Priors here can be controversial, depending on what literature we look to. Well-being is often left-skewed, so we could go for a skewed normal distribution for the model. However, that might be too strong an assumption, which is why I’ll use a model that assumes a normal Gaussian distribution.
As for the specific priors:
- When social media use is at zero between people and at zero within, I’ll simply assume a normal distribution centered on the midpoint of the scale (i.e., 3). As for the SD of that distribution, we know the bounds of the scale, so an SD of 1 will have 95% of cases within 1 and 5, which is exactly what we want.
- As for the slopes: There are two papers I know of that found a negative, small effect, but another paper that found a negligent one.
The larger literature on self-reported media use and well-being also finds very small negative effects, in the range of \(\beta\) = .05.
We’re on the unstandardized scale here.
So if we assume that well-being is at the midpoint of the scale (i.e., 3), the maximum effect an increase in social media time could have is to bring well-being to its ceiling or floor.
That would imply a standard deviation of 1 again - that depends on the scale of the predictor, though.
Right now, it’s in minutes and we’d need to scale the prior accordingly.
This paper found that with each increase of one standard deviation in social media time, well-being on a 7-point Likert-scale went down by -.06 units.
The standard deviation was 190.
Therefore (with some crude math), one hour of social media use was associated with
190/60 * -.06= -0.19 raw units on the outcome scale. From the literature we also know that massive (unstandardized) effects are rare if impossible. Therefore, I’ll center the slope distribution on a small negative value. To be conservative, I’ll take about 75% of the -.19 we found above (say -0.15 Likert-scales) with a somewhat tight standard deviation (say 0.3). This way, 95% of effects will be within -.75 (-.15 + (2 times 0.2)) and 0.45 Likert-points on the outcome scale. Note that we’re on the between-level here: a user with one more hour of social media use will report, on average, a .15 lower score on well-being than someone else with an hour less. - As for the within-effect, most research points toward negligible within-effects, and all of them are on self-reported media use. So here I’ll assume a slopes that varies around zero, but I’ll allow a wider standard deviation than for the between-effect, 0.4. That way, the average within-person effect will be zero and it assumes that 95% of effects are within -0.8 and + 0.8 Likert-points.
- For everything else, I’ll once more go with the default
brmspriors.
Let’s set those priors. Note that we’re still on the minute scale for the predictor, but I specified priors above for hours. That’s why I transform the social media variables to hours (and center) to make interpretation easier. That also helps us set the priors we specified above.
# create hour variables
dat <-
dat %>%
mutate(
across(
c(social_media_objective, social_media_subjective),
~ .x / 60, # divide by 60 to get hours
.names = "{col}_hours"
)
)
# center to get between and within variables
dat <-
dat %>%
group_by(id) %>%
mutate(
across(
ends_with("hours"),
list(
between = ~ mean(.x, na.rm = TRUE),
within = ~.x - mean(.x, na.rm = TRUE)
)
)
) %>%
ungroup()
# set priors
priors_model7 <-
c(
# intercept
prior(normal(3, 1), class = Intercept),
# slopes for between
prior(normal(-0.15, 0.30), class = b, coef = "social_media_objective_hours_between"),
# slopes for between
prior(normal(0, 0.40), class = b, coef = "social_media_objective_hours_within")
)Okay, let’s run the model.
model7 <-
brm(
data = dat,
family = gaussian,
prior = priors_model7,
well_being_state ~
1 +
social_media_objective_hours_between +
social_media_objective_hours_within +
(
1 +
social_media_objective_hours_within |
id
) +
(1 |day),
iter = 5000,
warmup = 2000,
chains = 4,
cores = 4,
seed = 42,
control = list(
adapt_delta = 0.99
),
file = here("models", "model7")
)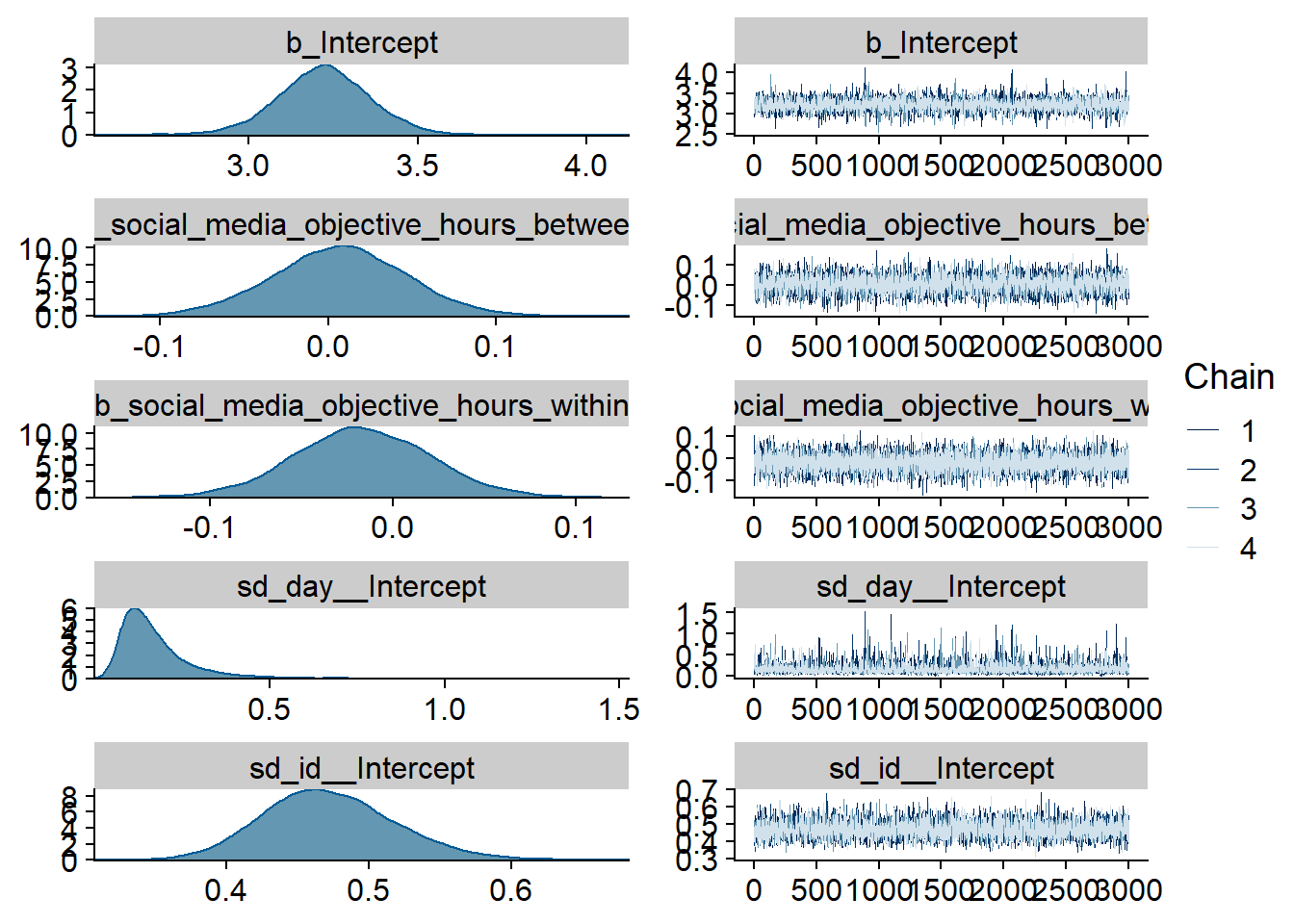
Figure 4.59: Traceplots and posterior distributions for Model 7
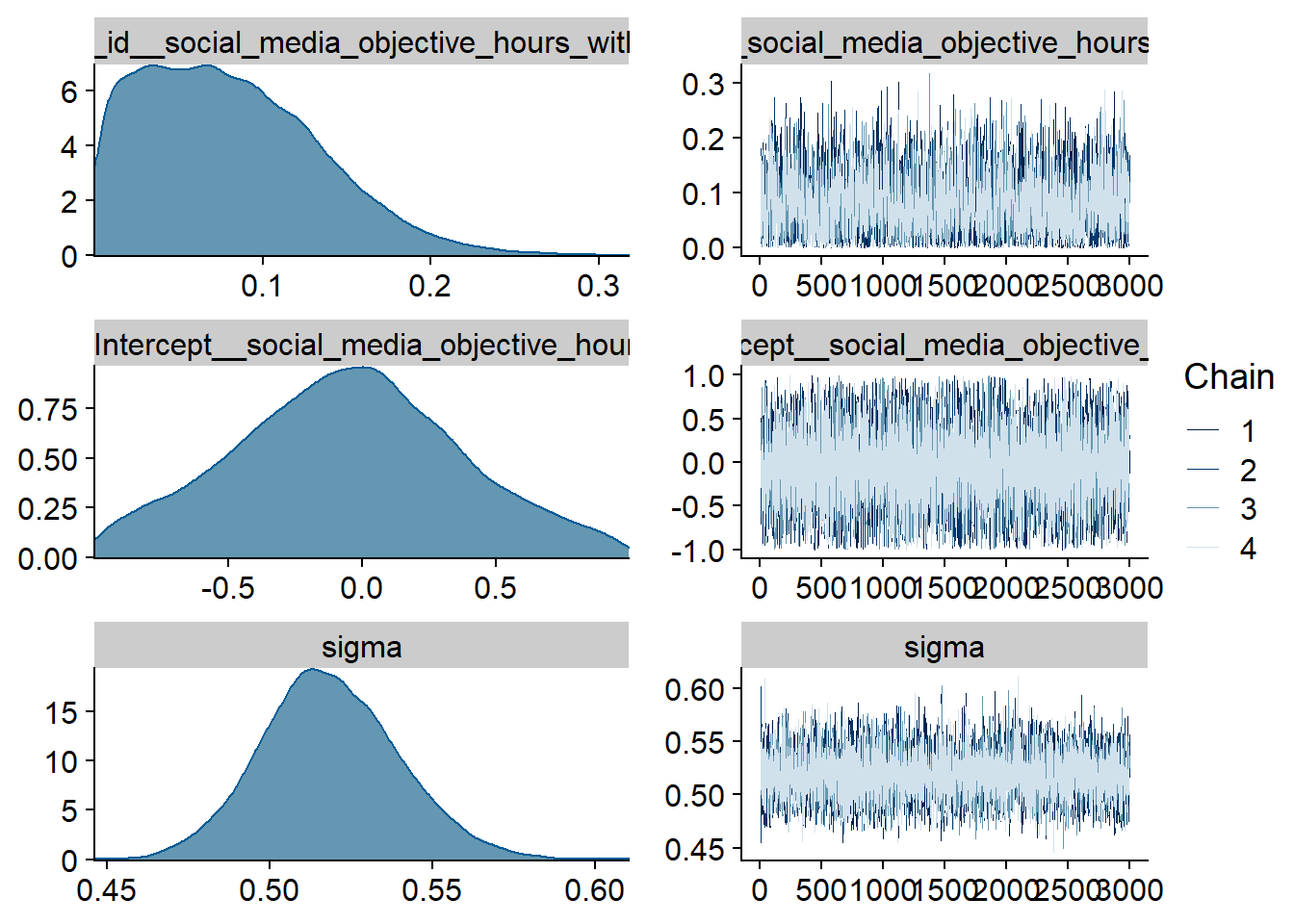
Figure 4.60: Traceplots and posterior distributions for Model 7
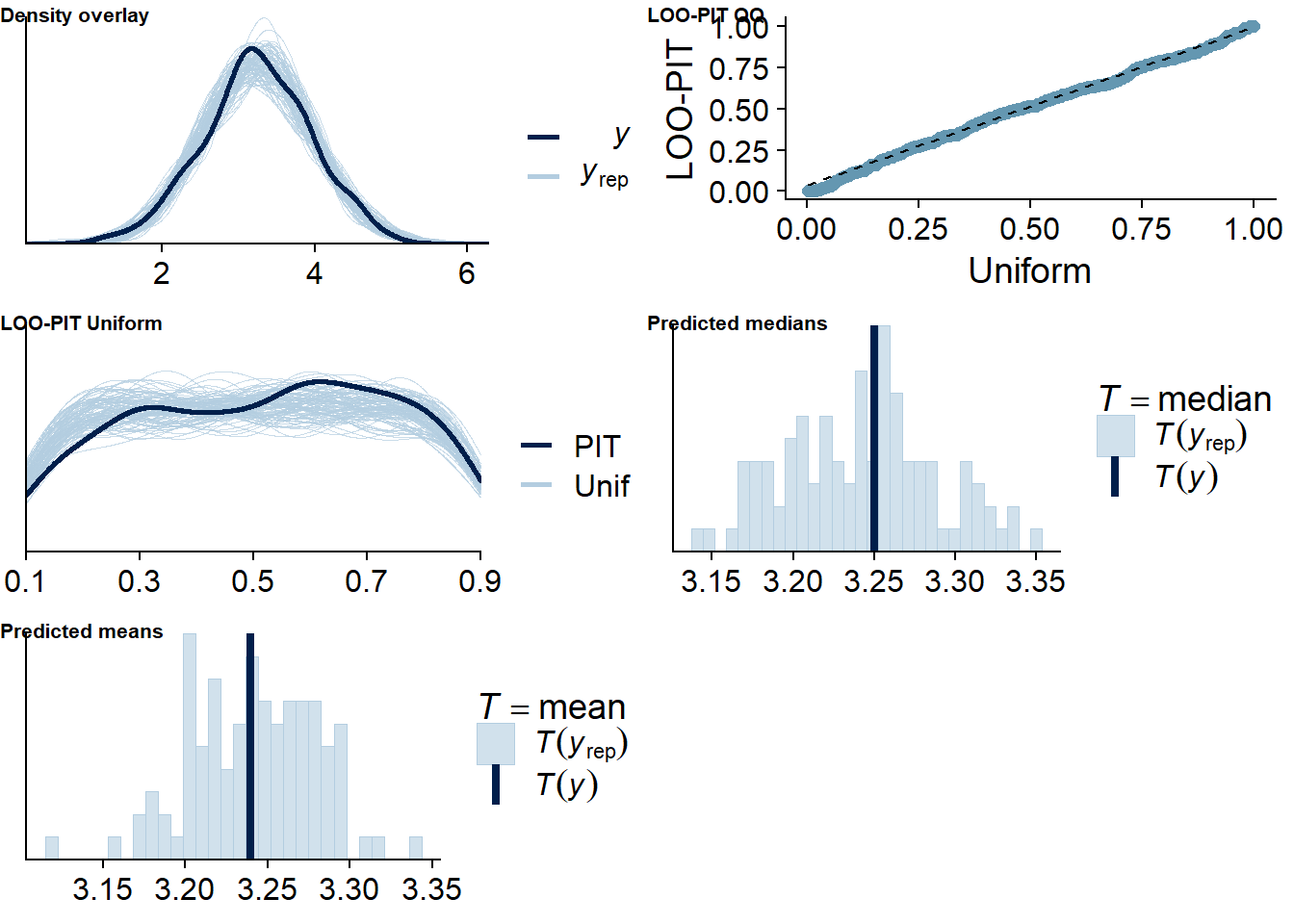
Figure 4.61: Posterior predictive checks for Model 7
One case is flagged as potentially influential, but unproblematic when calculated directly.
## 1 problematic observation(s) found.
## The model will be refit 1 times.##
## Fitting model 1 out of 1 (leaving out observation 65)## Start sampling##
## Computed from 12000 by 425 log-likelihood matrix
##
## Estimate SE
## elpd_loo -370.4 16.1
## p_loo 79.0 5.3
## looic 740.7 32.2
## ------
## Monte Carlo SE of elpd_loo is 0.1.
##
## Pareto k diagnostic values:
## Count Pct. Min. n_eff
## (-Inf, 0.5] (good) 410 96.5% 1038
## (0.5, 0.7] (ok) 15 3.5% 569
## (0.7, 1] (bad) 0 0.0% <NA>
## (1, Inf) (very bad) 0 0.0% <NA>
##
## All Pareto k estimates are ok (k < 0.7).
## See help('pareto-k-diagnostic') for details.Let’s inspect the summary. The relation is estimated to be extremely close to zero, so really no interpretative wiggle room there. I think this is pretty convincing evidence for the lack of an effect.
## Family: gaussian
## Links: mu = identity; sigma = identity
## Formula: well_being_state ~ 1 + social_media_objective_hours_between + social_media_objective_hours_within + (1 + social_media_objective_hours_within | id) + (1 | day)
## Data: study1 (Number of observations: 425)
## Samples: 4 chains, each with iter = 5000; warmup = 2000; thin = 1;
## total post-warmup samples = 12000
##
## Priors:
## b_social_media_objective_hours_between ~ normal(-0.15, 0.3)
## b_social_media_objective_hours_within ~ normal(0, 0.4)
## Intercept ~ normal(3, 1)
## L ~ lkj_corr_cholesky(1)
## sd ~ student_t(3, 0, 2.5)
## sigma ~ student_t(3, 0, 2.5)
##
## Group-Level Effects:
## ~day (Number of levels: 5)
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## sd(Intercept) 0.17 0.11 0.05 0.47 1.00 3575 5449
##
## ~id (Number of levels: 94)
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## sd(Intercept) 0.47 0.05 0.39 0.57 1.00 4536 7237
## sd(social_media_objective_hours_within) 0.08 0.05 0.00 0.20 1.00 3112 4958
## cor(Intercept,social_media_objective_hours_within) -0.05 0.42 -0.86 0.80 1.00 10858 6678
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## Intercept 3.22 0.14 2.94 3.50 1.00 4255 5282
## social_media_objective_hours_between 0.01 0.04 -0.07 0.08 1.00 5089 6626
## social_media_objective_hours_within -0.02 0.04 -0.09 0.06 1.00 15746 8839
##
## Family Specific Parameters:
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## sigma 0.52 0.02 0.48 0.56 1.00 10146 8819
##
## Samples were drawn using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).
Figure 4.62: Effects plot for Model 7
4.5.2 Model 8: Subjective use predicting well-being
Next, we’ll predict well-being on that day with subjective social media use on that day. There’s conflicting information in the literature: I’m aware of a paper that says subjective estimates overstate the relation, but of another that says the opposite. Therefore,
The specific priors:
- For the intercept, I’ll use the same prior as with Model 7.
- As for the slopes: This paper reports median effect sizes across all specifications of \(\beta\) = -.08, with effect sizes larger on the between-level, but small to negligible on the within-level. That’s among adolescents and for a longer time frame. This explicitly compared subjective to objective and found that subjective is a much stronger predictor (up to four times the effect size). This says subjective estimates ever so slightly understimate relations between use and well-being, though. Overall, I’d say we can expect a somewhat more negative between-person relation, which would lead us to believe a mean negative effect of -.20 Likert-points on well-being and 0.4 standard deviation to allow for more extreme slopes. That would bring 95% of effects within -1 and 0.6 Likert-points, which is rather generous for media effects.
- For the within-effect, I’ll stick to the same prior as for the previous model.
- For everything else, I’ll once more go with the default
brmspriors.
Let’s set those priors.
# set priors
priors_model8 <-
c(
# intercept
prior(normal(3, 1), class = Intercept),
# slopes for between
prior(normal(-0.2, 0.4), class = b, coef = "social_media_subjective_hours_between"),
# slopes for between
prior(normal(0, 0.40), class = b, coef = "social_media_subjective_hours_within")
)Okay, let’s run the model.
model8 <-
brm(
data = dat,
family = gaussian,
prior = priors_model8,
well_being_state ~
1 +
social_media_subjective_hours_between +
social_media_subjective_hours_within +
(
1 +
social_media_subjective_hours_within |
id
) +
(1 |day),
iter = 5000,
warmup = 2000,
chains = 4,
cores = 4,
seed = 42,
control = list(
adapt_delta = 0.99
),
file = here("models", "model8")
)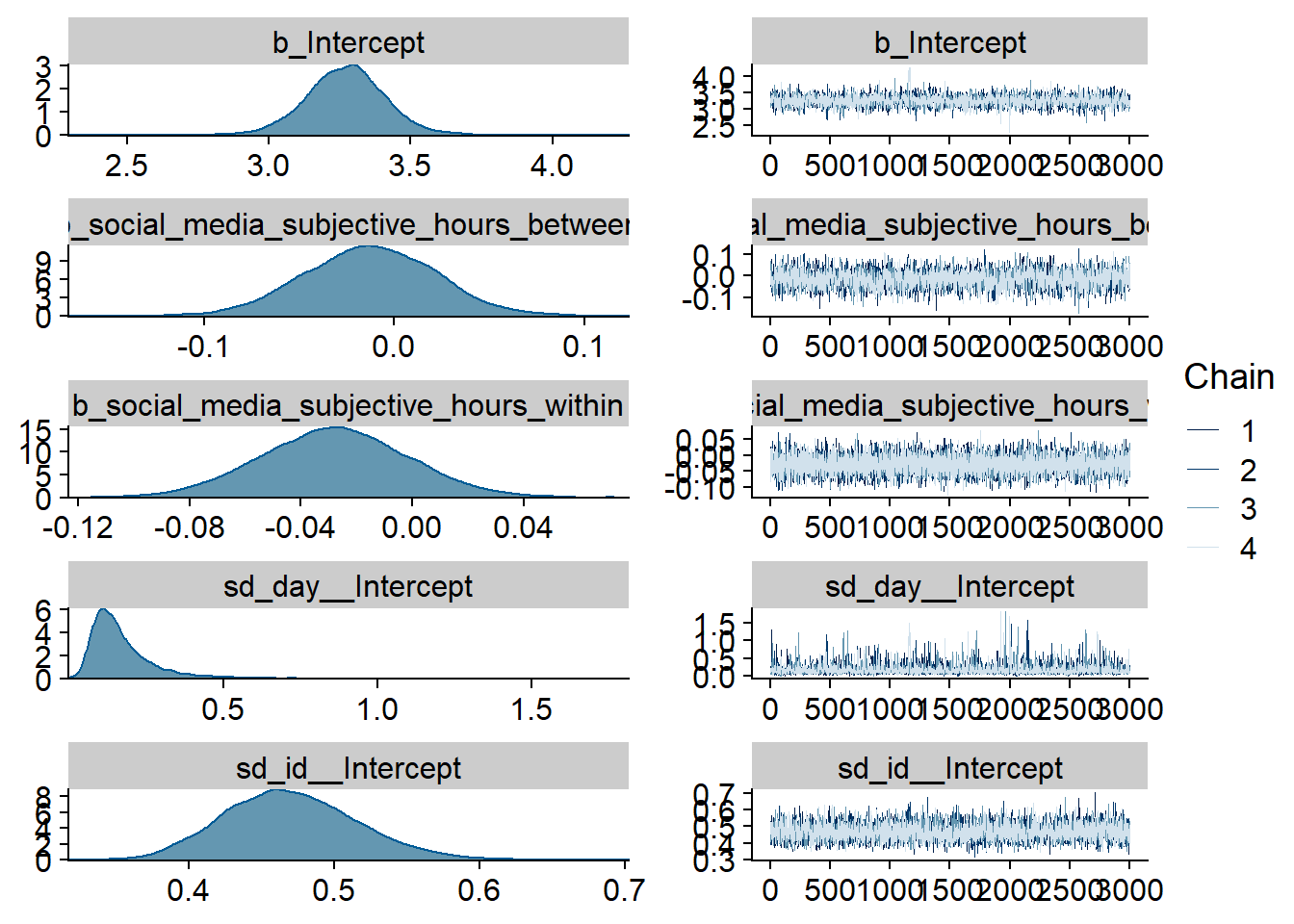
Figure 4.63: Traceplots and posterior distributions for Model 8
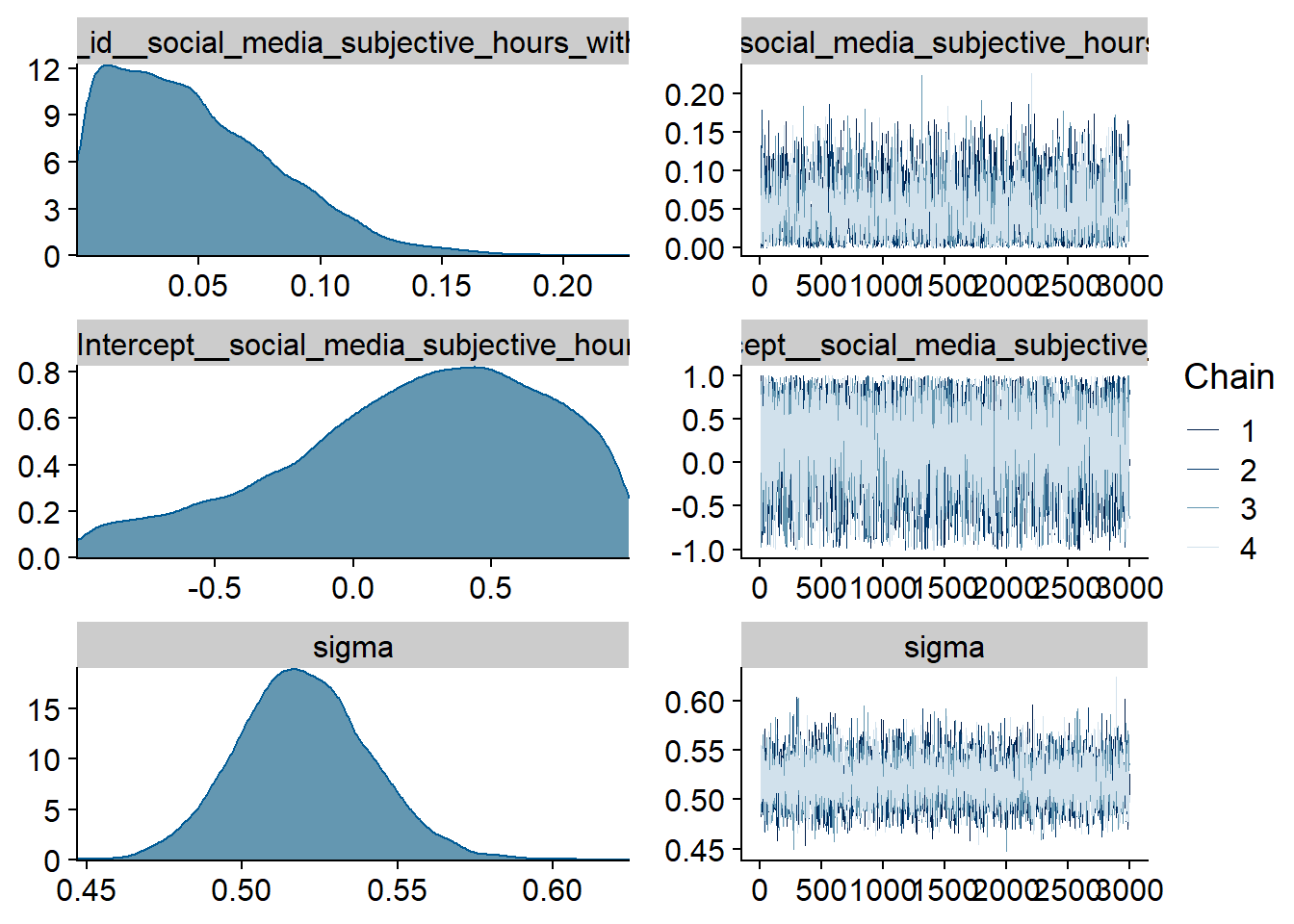
Figure 4.64: Traceplots and posterior distributions for Model 8
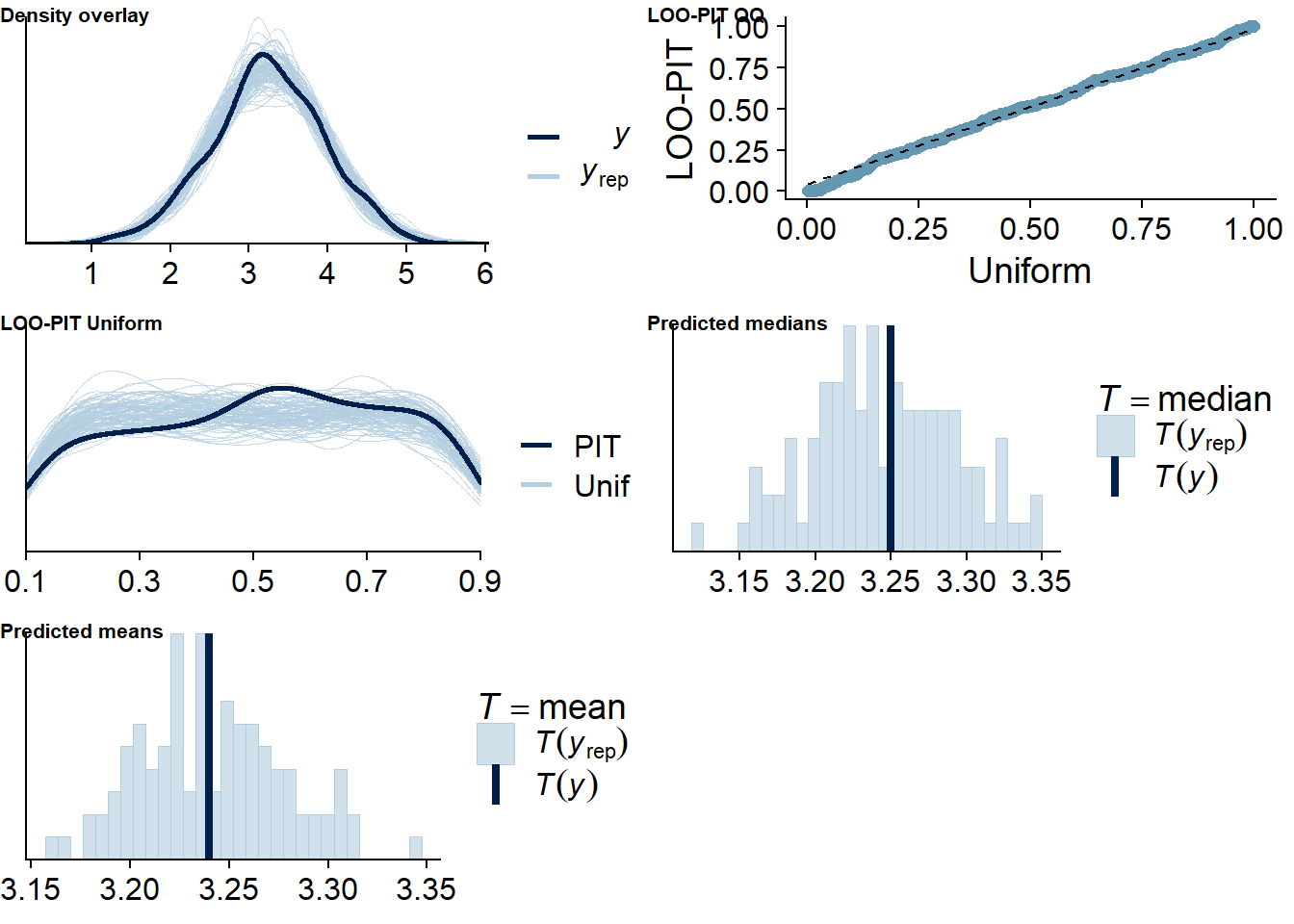
Figure 4.65: Posterior predictive checks for Model 8
One case is flagged as potentially influential, but unproblematic after being calculated directly..
## 1 problematic observation(s) found.
## The model will be refit 1 times.##
## Fitting model 1 out of 1 (leaving out observation 413)## Start sampling##
## Computed from 12000 by 425 log-likelihood matrix
##
## Estimate SE
## elpd_loo -371.1 16.2
## p_loo 78.7 5.4
## looic 742.2 32.4
## ------
## Monte Carlo SE of elpd_loo is 0.2.
##
## Pareto k diagnostic values:
## Count Pct. Min. n_eff
## (-Inf, 0.5] (good) 405 95.3% 389
## (0.5, 0.7] (ok) 20 4.7% 554
## (0.7, 1] (bad) 0 0.0% <NA>
## (1, Inf) (very bad) 0 0.0% <NA>
##
## All Pareto k estimates are ok (k < 0.7).
## See help('pareto-k-diagnostic') for details.Let’s inspect the summary. The within effect is estimated to be negative, but extremely small. Also, the 95% CI includes zero, so we can’t be sure the effect is meaningful.
## Family: gaussian
## Links: mu = identity; sigma = identity
## Formula: well_being_state ~ 1 + social_media_subjective_hours_between + social_media_subjective_hours_within + (1 + social_media_subjective_hours_within | id) + (1 | day)
## Data: study1 (Number of observations: 425)
## Samples: 4 chains, each with iter = 5000; warmup = 2000; thin = 1;
## total post-warmup samples = 12000
##
## Priors:
## b_social_media_subjective_hours_between ~ normal(-0.2, 0.4)
## b_social_media_subjective_hours_within ~ normal(0, 0.4)
## Intercept ~ normal(3, 1)
## L ~ lkj_corr_cholesky(1)
## sd ~ student_t(3, 0, 2.5)
## sigma ~ student_t(3, 0, 2.5)
##
## Group-Level Effects:
## ~day (Number of levels: 5)
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## sd(Intercept) 0.18 0.13 0.05 0.51 1.00 2790 3833
##
## ~id (Number of levels: 94)
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## sd(Intercept) 0.47 0.05 0.39 0.57 1.00 3971 6987
## sd(social_media_subjective_hours_within) 0.05 0.03 0.00 0.13 1.00 3070 4785
## cor(Intercept,social_media_subjective_hours_within) 0.23 0.47 -0.83 0.95 1.00 8042 5984
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## Intercept 3.27 0.14 2.99 3.55 1.00 3189 4864
## social_media_subjective_hours_between -0.01 0.04 -0.08 0.06 1.00 3174 5245
## social_media_subjective_hours_within -0.03 0.03 -0.08 0.02 1.00 12266 8541
##
## Family Specific Parameters:
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## sigma 0.52 0.02 0.48 0.56 1.00 9007 8503
##
## Samples were drawn using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).
Figure 4.66: Effects plot for Model 8
4.5.3 Model 9: Accuracy predicting well-being
For this model, we ask whether the error people make in their estimation is related to their well-being on the state level. As for as I know, there’s little prior knowledge we could build on for the relation between accuracy and well-being.
As for the specific priors:
- Like before, at perfect accuracy (0% error) and zero deviation I’ll assume a normal distribution centered on the midpoint of the scale (i.e., 3) with the intercept. I’ll set the SD of that prior distribution to 1 once more.
- As for the slopes: There’s one paper that correlated the absolute discrepancy between subjective and objective social media use and well-being indicators aggregated over a week.
They found a \(\beta\) = .16 between log-transformed discrepancies and depression scores.
Discrepancy is a different measurement than the error that we calculated for accuracy.
Therefore, I’ll use a prior that’s centered on a negative relationship with a small SD.
Like above, I don’t expect that there will be large effects just based on the literature.
So I’ll assume a small effect (say -0.20 Likert-scales) with a somewhat wider standard deviation than in previous models (say 0.4).
This way, 95% of effects will be within -1 (-.20 + (2 times 0.3)) and 0.60 Likert-points on the outcome scale.
However,
erroris in percent, so we wouldn’t expect -.20 Likert-points with every one-percent increase. Rather, I’d say the above effect is easier to specify for a standard deviation change inerror, which is why I standardizeerror: one standard deviation increase in accuracy will be associated with, on average, a .20 lower score on well-being on the between level. - I have no information for the within-level, which is why I use a prior centered on zero with the same wide standard deviation.
- For everything else, I’ll once more go with the default
brmspriors.
Let’s set those priors and center at the same time.
# standardize error
dat <-
dat %>%
mutate(
error_s = scale(error, center = FALSE, scale = TRUE)
)
# center to get between and within
dat <-
dat %>%
group_by(id) %>%
mutate(
across(
error_s,
list(
between = ~ mean(.x, na.rm = TRUE),
within = ~.x - mean(.x, na.rm = TRUE)
)
)
) %>%
ungroup()
# set priors
priors_model9 <-
c(
# intercept
prior(normal(3, 1), class = Intercept),
# slopes for between
prior(normal(-0.2, 0.4), class = b, coef = "error_s_between"),
# slopes for between
prior(normal(0, 0.40), class = b, coef = "error_s_within")
)And run the model.
model9 <-
brm(
data = dat,
family = gaussian,
prior = priors_model9,
well_being_state ~
1 +
error_s_between +
error_s_within +
(
1 +
error_s_within |
id
) +
(1 |day),
iter = 5000,
warmup = 2000,
chains = 4,
cores = 4,
seed = 42,
control = list(
adapt_delta = 0.999
),
file = here("models", "model9")
)
Figure 4.67: Traceplots and posterior distributions for Model 9
Figure 4.68: Traceplots and posterior distributions for Model 9
Figure 4.69: Posterior predictive checks for Model 9
Six cases are flagged as potentially influential, which is why we calculate ELPD directly.
## 6 problematic observation(s) found.
## The model will be refit 6 times.##
## Fitting model 1 out of 6 (leaving out observation 23)##
## Fitting model 2 out of 6 (leaving out observation 24)##
## Fitting model 3 out of 6 (leaving out observation 255)##
## Fitting model 4 out of 6 (leaving out observation 289)##
## Fitting model 5 out of 6 (leaving out observation 397)##
## Fitting model 6 out of 6 (leaving out observation 417)## Start sampling
## Start sampling
## Start sampling
## Start sampling
## Start sampling
## Start sampling##
## Computed from 12000 by 423 log-likelihood matrix
##
## Estimate SE
## elpd_loo -367.8 16.4
## p_loo 81.4 6.0
## looic 735.6 32.8
## ------
## Monte Carlo SE of elpd_loo is 0.2.
##
## Pareto k diagnostic values:
## Count Pct. Min. n_eff
## (-Inf, 0.5] (good) 407 96.2% 47
## (0.5, 0.7] (ok) 16 3.8% 492
## (0.7, 1] (bad) 0 0.0% <NA>
## (1, Inf) (very bad) 0 0.0% <NA>
##
## All Pareto k estimates are ok (k < 0.7).
## See help('pareto-k-diagnostic') for details.Let’s inspect the summary. The within effect is estimated to be negative, but really small. Also, the 95% CI includes zero and quite wide, so we can’t be sure the effect is meaningful.
## Family: gaussian
## Links: mu = identity; sigma = identity
## Formula: well_being_state ~ 1 + error_s_between + error_s_within + (1 + error_s_within | id) + (1 | day)
## Data: study1 (Number of observations: 423)
## Samples: 4 chains, each with iter = 5000; warmup = 2000; thin = 1;
## total post-warmup samples = 12000
##
## Priors:
## b_error_s_between ~ normal(-0.2, 0.4)
## b_error_s_within ~ normal(0, 0.4)
## Intercept ~ normal(3, 1)
## L ~ lkj_corr_cholesky(1)
## sd ~ student_t(3, 0, 2.5)
## sigma ~ student_t(3, 0, 2.5)
##
## Group-Level Effects:
## ~day (Number of levels: 5)
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## sd(Intercept) 0.19 0.15 0.06 0.53 1.00 3196 3812
##
## ~id (Number of levels: 94)
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## sd(Intercept) 0.47 0.04 0.39 0.56 1.00 4652 8200
## sd(error_s_within) 0.14 0.09 0.01 0.34 1.00 2730 5295
## cor(Intercept,error_s_within) 0.25 0.42 -0.72 0.93 1.00 9691 7015
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## Intercept 3.25 0.13 3.01 3.48 1.00 4066 3419
## error_s_between -0.02 0.07 -0.16 0.12 1.00 6313 7487
## error_s_within -0.03 0.06 -0.15 0.08 1.00 13197 8720
##
## Family Specific Parameters:
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## sigma 0.51 0.02 0.48 0.56 1.00 9411 9387
##
## Samples were drawn using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).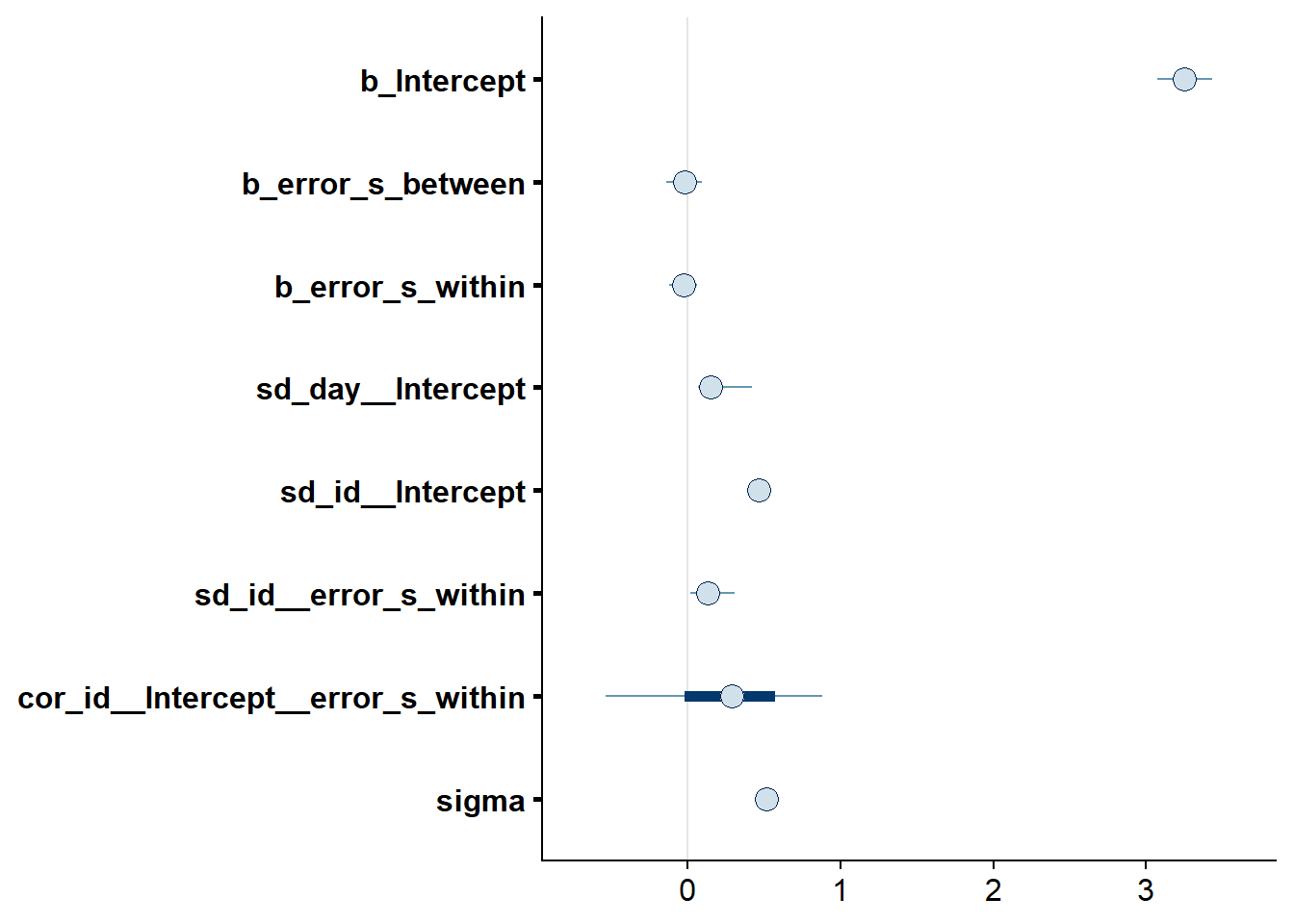
Figure 4.70: Effects plot for Model 9
4.6 Relation between subjective and objective use
Something that wasn’t part of the original plan: inspecting the relation between subjective and objective use a bit closer. The absolute difference between the two wasn’t that large, so it’s possible that the difference isn’t meaningfully different from zero. For that reason, I predict subjective use with objective use and then inspect the parameter. The intercept will tell us whether the subjective overestimate is different from zero (so an indicator of bias). The slope will tell us how sensitive subjective is to the true value. And the sigma will tell us how much error there is.
Unfortunately, the model has huge problems with a Gamma distribution as outcome, so I went for a normal distribution with the predictor grand-mean centered instead.
Also, I’m not using informed priors here and rely on the default brms priors.
For this question, I’m not interested in distinguishing between- and within-person effects.
model10 <-
brm(
data = dat,
family = gaussian,
social_media_subjective ~
1 +
social_media_objective +
(1 + social_media_objective | id) +
(1 | day),
iter = 5000,
warmup = 2000,
chains = 4,
cores = 4,
seed = 42,
control = list(adapt_delta = 0.99),
file = here("models", "model10.rds")
)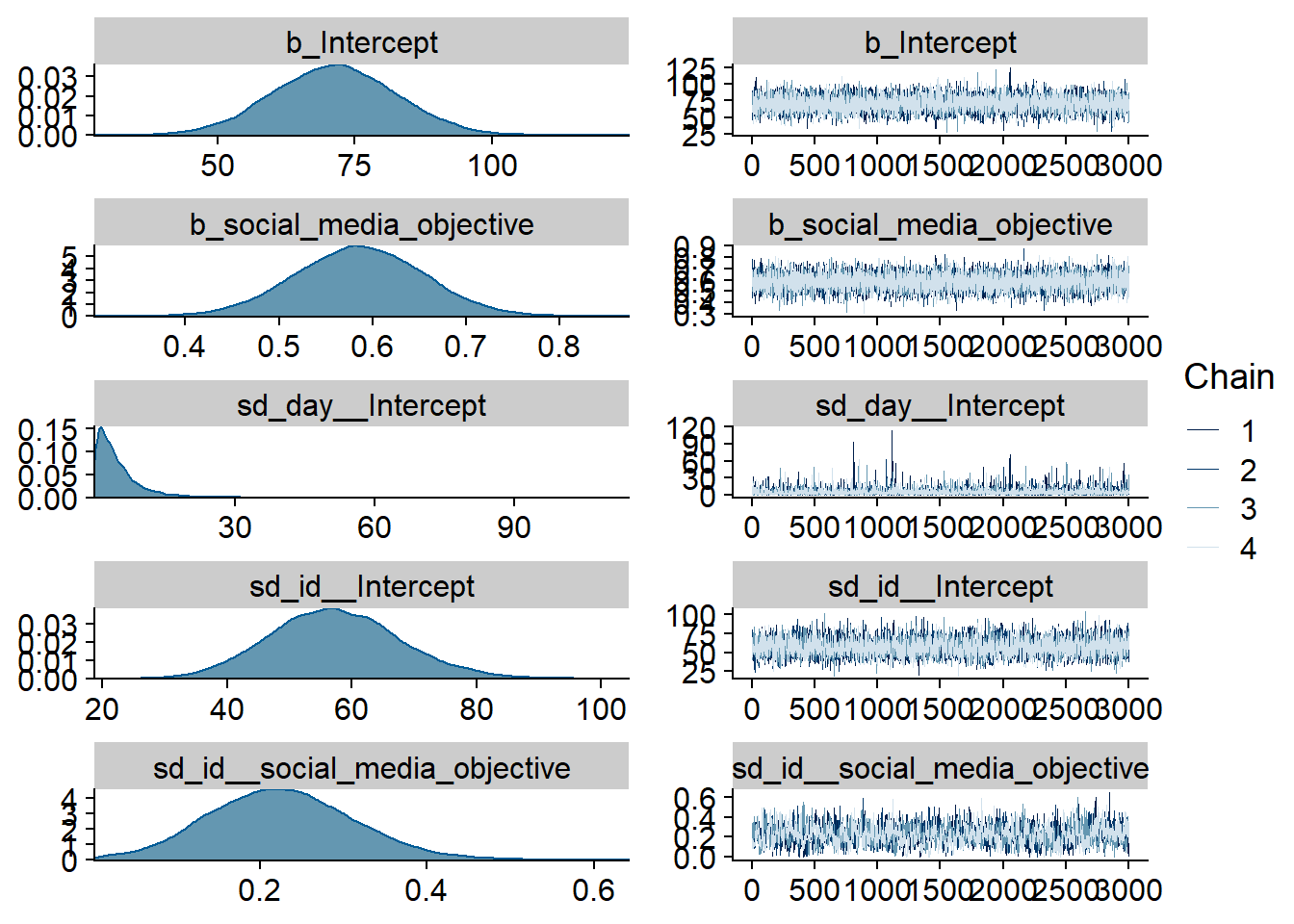
Figure 4.71: Traceplots and posterior distributions for Model 10
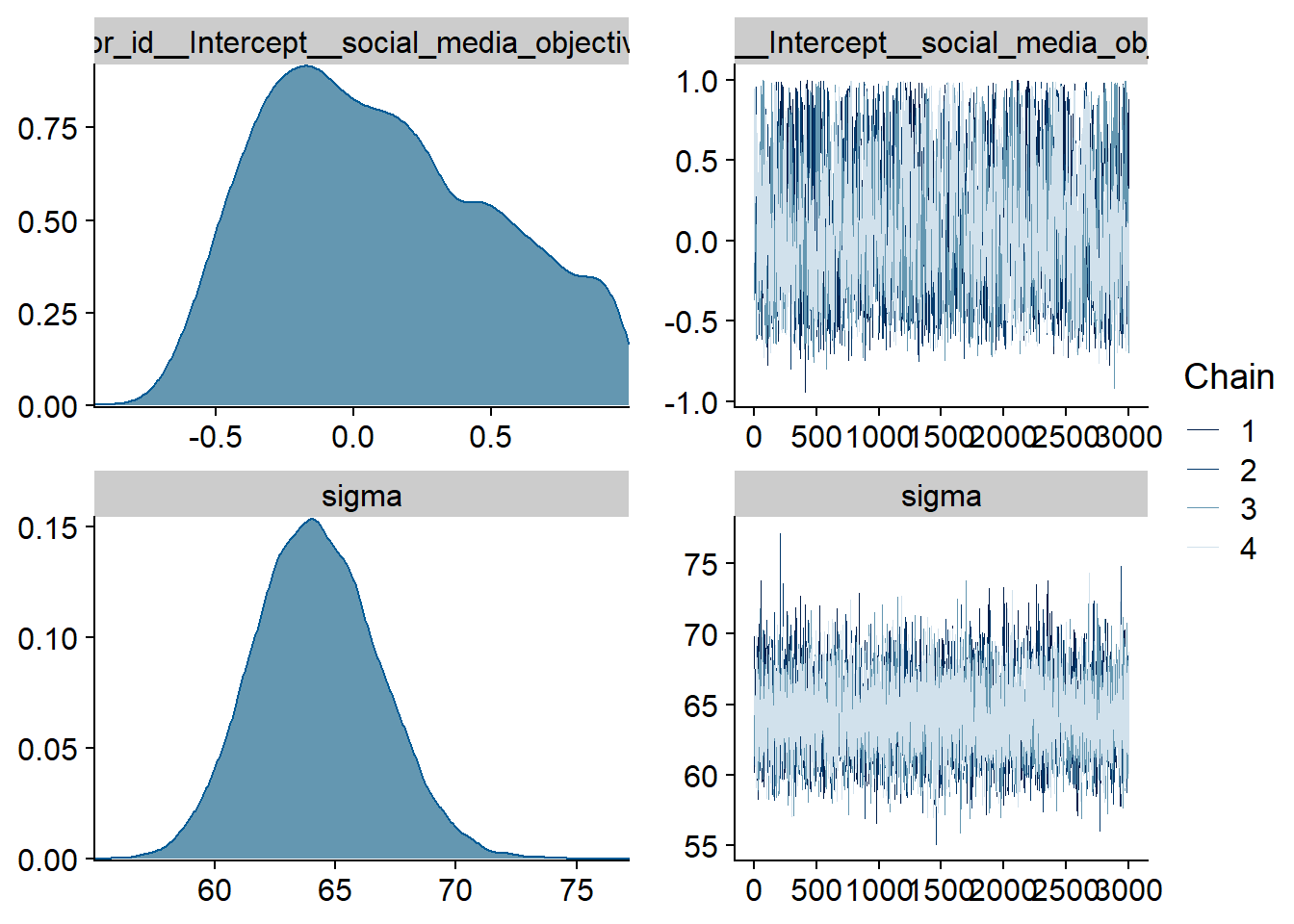
Figure 4.72: Traceplots and posterior distributions for Model 10
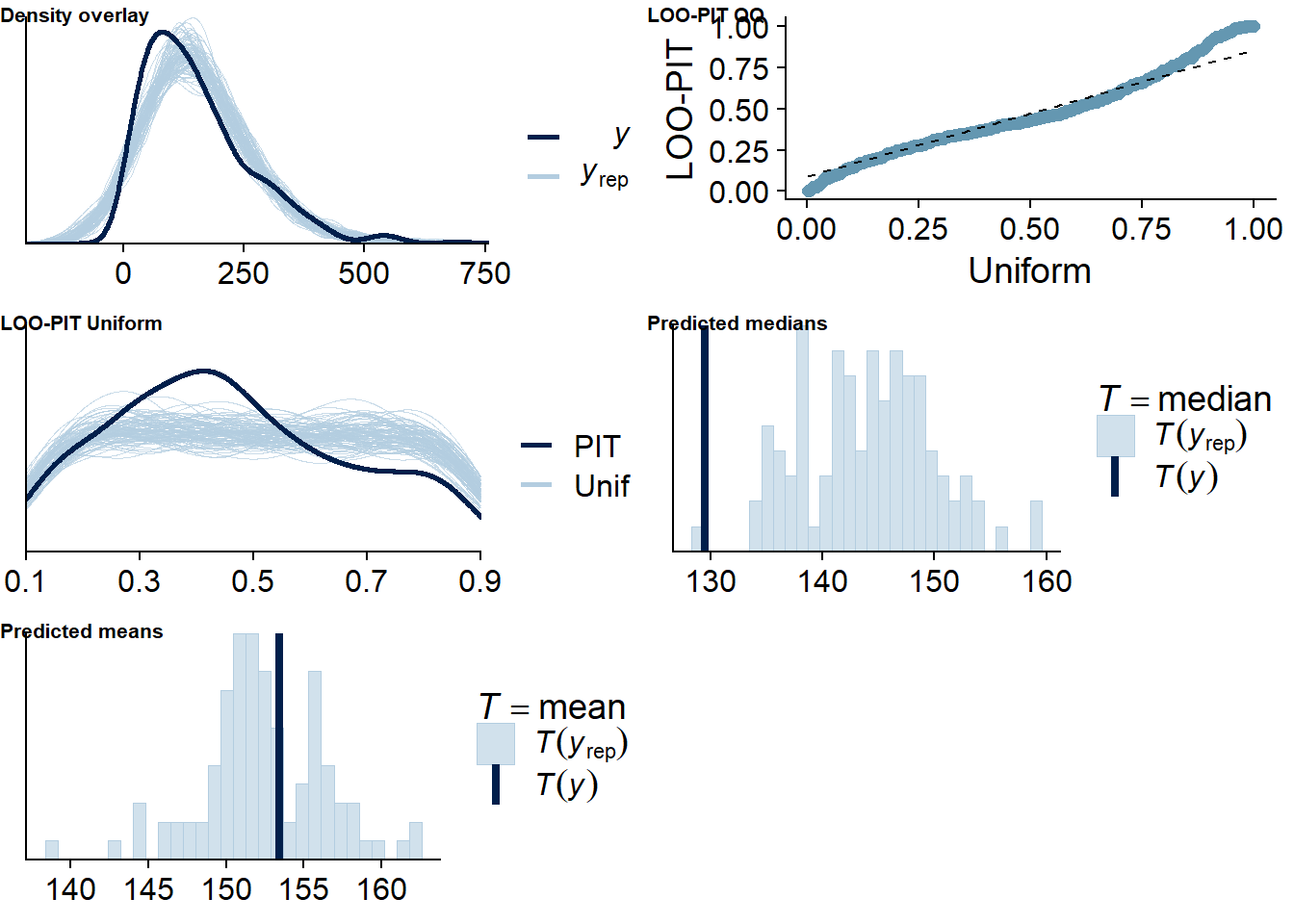
Figure 4.73: Posterior predictive checks for Model 10
Five cases are flagged as potentially influential, which is why we calculate ELPD directly.
## 5 problematic observation(s) found.
## The model will be refit 5 times.##
## Fitting model 1 out of 5 (leaving out observation 23)##
## Fitting model 2 out of 5 (leaving out observation 63)##
## Fitting model 3 out of 5 (leaving out observation 134)##
## Fitting model 4 out of 5 (leaving out observation 350)##
## Fitting model 5 out of 5 (leaving out observation 392)## Start sampling
## Start sampling
## Start sampling
## Start sampling
## Start sampling##
## Computed from 12000 by 428 log-likelihood matrix
##
## Estimate SE
## elpd_loo -2439.9 23.7
## p_loo 85.1 8.2
## looic 4879.8 47.4
## ------
## Monte Carlo SE of elpd_loo is 0.3.
##
## Pareto k diagnostic values:
## Count Pct. Min. n_eff
## (-Inf, 0.5] (good) 401 93.7% 39
## (0.5, 0.7] (ok) 27 6.3% 134
## (0.7, 1] (bad) 0 0.0% <NA>
## (1, Inf) (very bad) 0 0.0% <NA>
##
## All Pareto k estimates are ok (k < 0.7).
## See help('pareto-k-diagnostic') for details.Let’s inspect the summary: indeed the intercept is different from zero, which speaks for a bias/systematic error in overestimates of social media time.
## Family: gaussian
## Links: mu = identity; sigma = identity
## Formula: social_media_subjective ~ 1 + social_media_objective + (1 + social_media_objective | id) + (1 | day)
## Data: study1 (Number of observations: 428)
## Samples: 4 chains, each with iter = 5000; warmup = 2000; thin = 1;
## total post-warmup samples = 12000
##
## Priors:
## Intercept ~ student_t(3, 129.5, 103)
## L ~ lkj_corr_cholesky(1)
## sd ~ student_t(3, 0, 103)
## sigma ~ student_t(3, 0, 103)
##
## Group-Level Effects:
## ~day (Number of levels: 5)
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## sd(Intercept) 4.99 5.52 0.15 18.65 1.00 6023 6603
##
## ~id (Number of levels: 94)
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## sd(Intercept) 57.35 10.50 37.79 79.11 1.00 4648 7538
## sd(social_media_objective) 0.22 0.09 0.06 0.40 1.00 960 2260
## cor(Intercept,social_media_objective) 0.10 0.42 -0.57 0.93 1.00 1411 2450
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## Intercept 71.21 11.16 49.54 92.95 1.00 8331 9048
## social_media_objective 0.59 0.07 0.45 0.72 1.00 10561 8913
##
## Family Specific Parameters:
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## sigma 64.24 2.57 59.45 69.44 1.00 7416 8846
##
## Samples were drawn using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).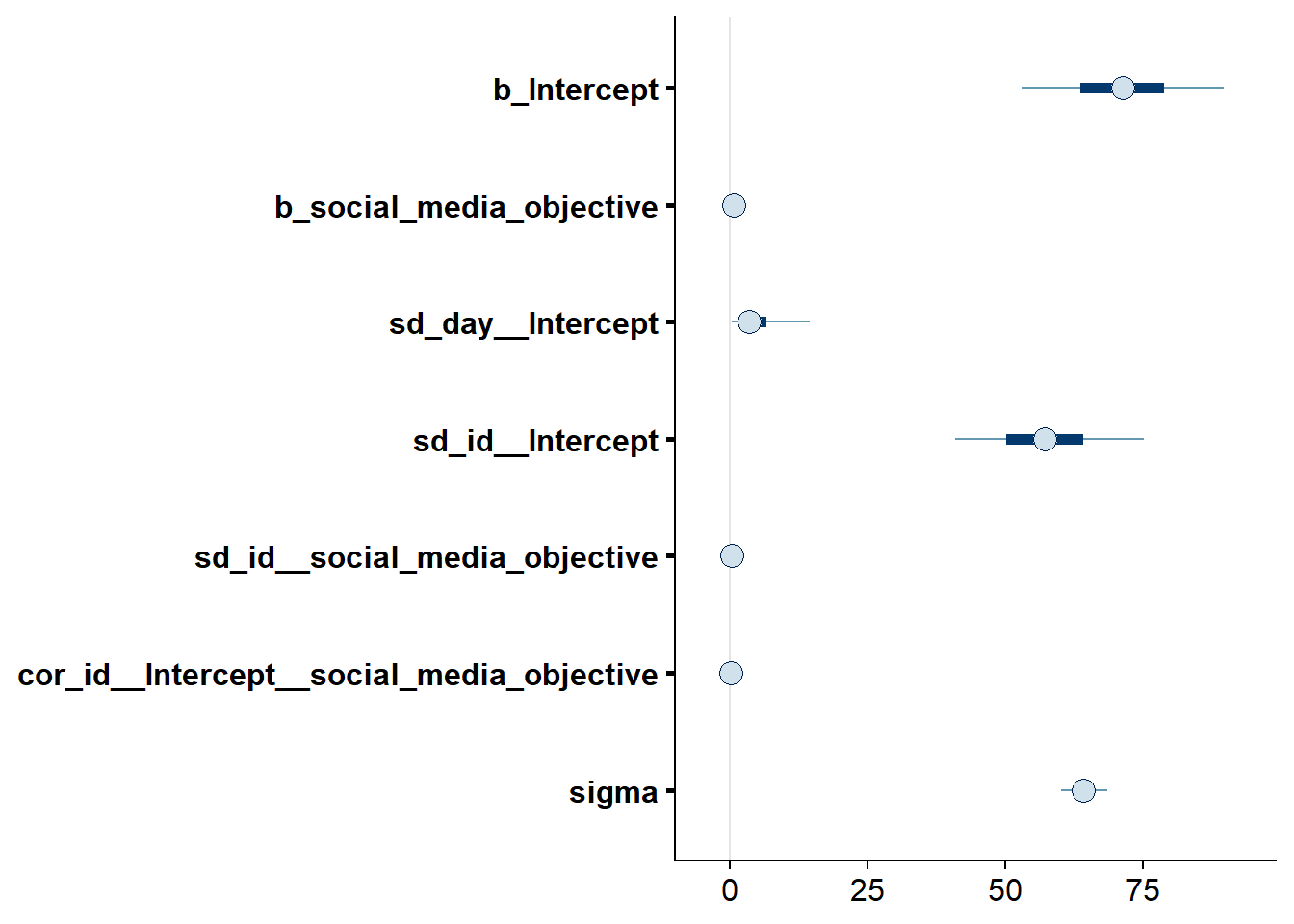
Figure 4.74: Effects plot for Model 10