4 Describe & explore
Here, we get the descriptive info for both studies that we report in the Method section of the paper.
We used these packages:
library(pacman)
p_load(
knitr,
here,
visdat,
scales,
patchwork,
sjPlot,
cowplot,
janitor,
ggbeeswarm,
lemon,
lubridate,
tidyverse
)Then, we load the data sets.
ac <- read_rds(here("data/noa/ac-excluded.rds"))
pvz <- read_rds(here("data/ea/pvz-excluded.rds"))
ac_full <- read_rds(here("data/noa/ac.rds"))
pvz_full <- read_rds(here("data/ea/pvz.rds"))Join the two data sets.
ac <- ac %>%
select(
player_id, gender, created, age,
spane_balance, spane_positive, spane_negative,
spane_game_balance, within_estimate, between_estimate,
autonomy,
competence, relatedness, enjoyment,
extrinsic, active_play, Hours
)
pvz <- pvz %>%
select(
player_id, gender, created = date, age,
spane_balance, spane_positive, spane_negative,
spane_game_balance, within_estimate, between_estimate,
autonomy,
competence, relatedness, enjoyment,
extrinsic, active_play, Hours
)
dat <- bind_rows(pvz, ac, .id = "Game") %>%
mutate(Game = factor(Game, labels = c("PvZ", "AC:NH")))4.1 Demographics
In total, 6484 players responded to the survey. Their mean age was M = 31 with a standard deviation of SD = 10.
Ages by study:
dat %>%
group_by(Game) %>%
summarise(
m_age = mean(age, na.rm = TRUE),
s_age = sd(age, na.rm = TRUE),
min_age = min(age, na.rm = TRUE),
max_age = max(age, na.rm = TRUE)
)
#> # A tibble: 2 x 5
#> Game m_age s_age min_age max_age
#> <fct> <dbl> <dbl> <dbl+lbl> <dbl+lbl>
#> 1 PvZ 34.9 11.8 18 99
#> 2 AC:NH 30.9 9.68 18 99Then let’s look at the sex distribution.
tabyl(pvz, gender) %>% adorn_pct_formatting()
#> gender n percent
#> Male 404 78.1%
#> Female 94 18.2%
#> Other 2 0.4%
#> Prefer not to say 17 3.3%
tabyl(ac, gender) %>% adorn_pct_formatting()
#> gender n percent valid_percent
#> Female 2462 41.3% 42.3%
#> Male 3124 52.4% 53.6%
#> Other 153 2.6% 2.6%
#> Prefer not to say 88 1.5% 1.5%
#> Item was never rendered for this user. 0 0.0% 0.0%
#> <NA> 140 2.3% -4.2 Survey dates
dat %>%
mutate(Date = as.Date(created)) %>%
count(Game, Date) %>%
kable()| Game | Date | n |
|---|---|---|
| PvZ | 2020-08-11 | 69 |
| PvZ | 2020-08-12 | 9 |
| PvZ | 2020-08-13 | 7 |
| PvZ | 2020-08-15 | 1 |
| PvZ | 2020-08-16 | 1 |
| PvZ | 2020-08-17 | 1 |
| PvZ | 2020-08-18 | 1 |
| PvZ | 2020-08-20 | 1 |
| PvZ | 2020-08-21 | 1 |
| PvZ | 2020-08-23 | 1 |
| PvZ | 2020-08-28 | 1 |
| PvZ | 2020-09-24 | 320 |
| PvZ | 2020-09-25 | 64 |
| PvZ | 2020-09-26 | 21 |
| PvZ | 2020-09-27 | 12 |
| PvZ | 2020-09-28 | 5 |
| PvZ | 2020-09-29 | 2 |
| AC:NH | 2020-10-27 | 236 |
| AC:NH | 2020-10-28 | 4942 |
| AC:NH | 2020-10-29 | 450 |
| AC:NH | 2020-10-30 | 171 |
| AC:NH | 2020-10-31 | 57 |
| AC:NH | 2020-11-01 | 55 |
| AC:NH | 2020-11-02 | 56 |
4.3 Response rates
# PvZ wave 1
p1 <- nrow(pvz_full %>% filter(date < ymd("2020-09-01")))
# PvZ wave 2
p2 <- nrow(pvz_full %>% filter(date > ymd("2020-09-01")))
# AC:NH
a1 <- nrow(ac)
tibble(
Game = c("PvZ w1", "PvZ w2", "AC:NH"),
Invitations = c(50000, 200000, 342825),
Responses = c(p1, p2, a1),
Rate = percent(Responses/Invitations, .01)
)
#> # A tibble: 3 x 4
#> Game Invitations Responses Rate
#> <chr> <dbl> <int> <chr>
#> 1 PvZ w1 50000 94 0.19%
#> 2 PvZ w2 200000 424 0.21%
#> 3 AC:NH 342825 5967 1.74%How many individuals had telemetry
foo <- function(data) {
data %>%
transmute(has_telemetry = !is.na(Hours)) %>%
tabyl(has_telemetry) %>%
adorn_pct_formatting()
}
map(list(pvz=pvz_full, ac=ac_full), foo) %>% kable
|
|
4.4 Missingness
vis_miss(pvz)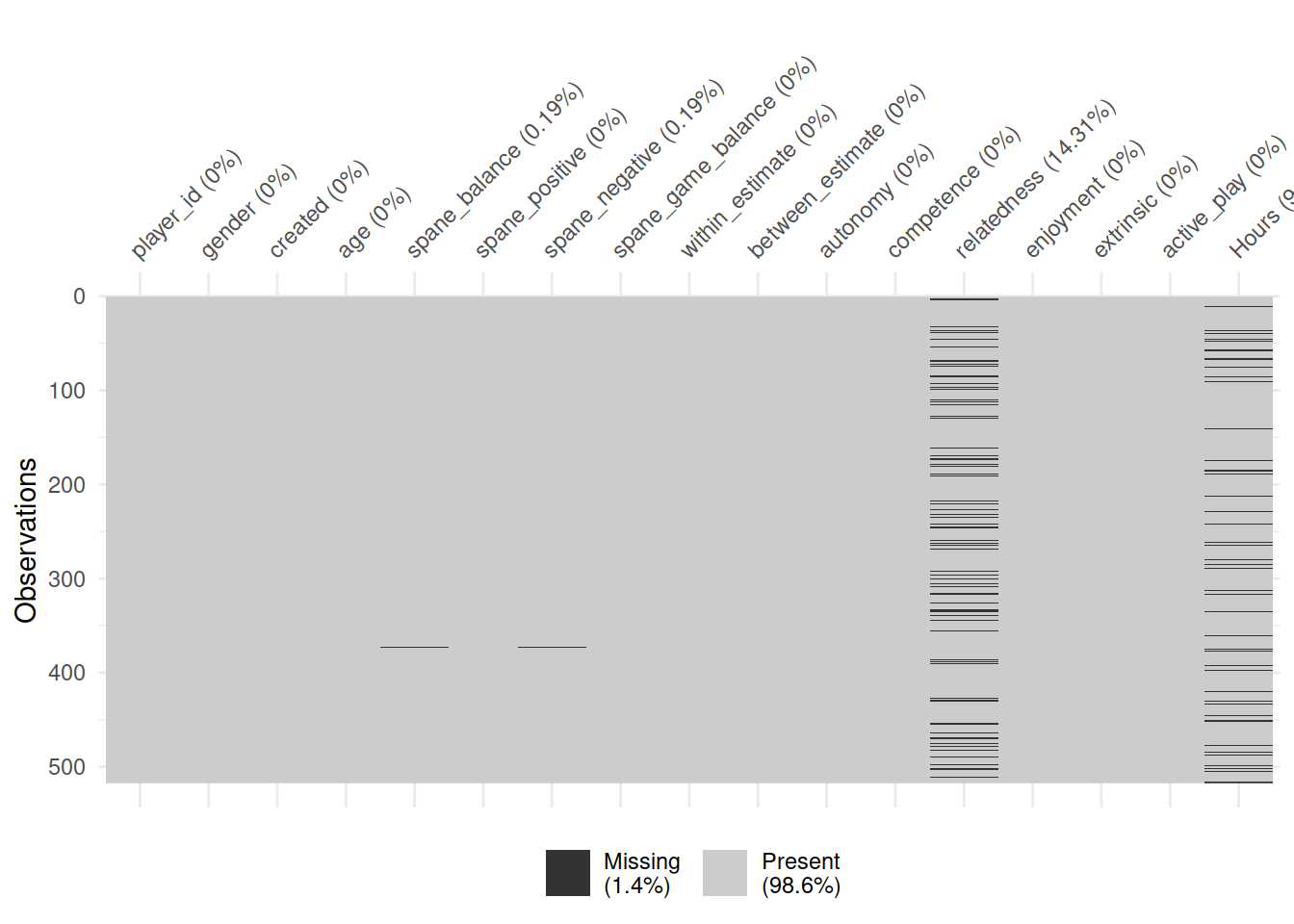
vis_miss(ac)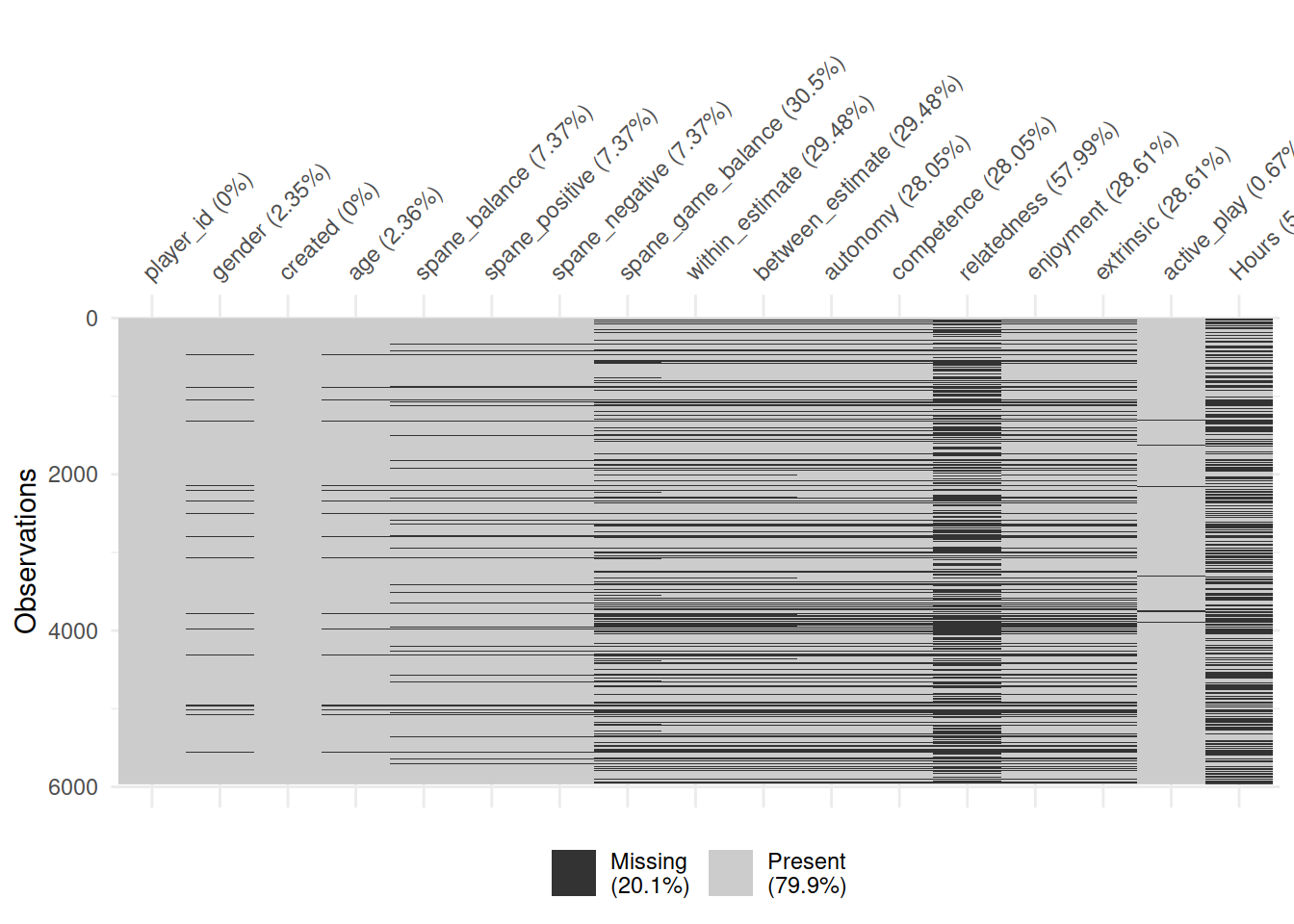
4.5 Univariate figures
This figure shows the distribution of well-being and motivation scores for both studies
tmp <- dat %>%
pivot_longer(c(spane_balance, autonomy:extrinsic))
tmp <- tmp %>%
mutate(
name = factor(
name,
levels = c(
"spane_balance", "extrinsic", "enjoyment",
"autonomy", "competence", "relatedness"
),
labels = c(
"Player well-being", "Extrinsic motivation", "Intrinsic motivation",
"Player autonomy", "Player competence", "Player relatedness"
)
)
)
tmp2 <- tmp %>%
group_by(Game, name) %>%
summarise(value = mean(value, na.rm = TRUE))
filter(tmp, Game == "PvZ") %>%
ggplot(aes(value, col = Game, fill = Game)) +
coord_cartesian(ylim = c(-1, 1)) +
scale_color_manual(
values = rev(colors), aesthetics = c("color", "fill"),
guide = guide_legend(reverse = TRUE)
) +
geom_histogram(
aes(y = stat(ncount)),
col = "white", bins = 20
) +
geom_histogram(
data = filter(tmp, Game == "AC:NH"),
aes(y = stat(ncount)*-1), col = "white", bins = 20
) +
scale_y_continuous(
breaks = c(-1, -.5, 0, .5, 1), labels = c(1, .5, 0, .5, 1)
) +
scale_x_continuous("Value", breaks = pretty_breaks()) +
geom_point(
data = tmp2, aes(y = -1.01),
shape = 25, size = 2.5
) +
# Make sure 1 is shown on all figures by drawing a blank geom
geom_blank(data = tibble(Game = "PvZ", value = 1)) +
labs(y = "Normalized count") +
facet_wrap("name", scales = "free") +
theme(axis.title.x = element_blank(), legend.position = "right")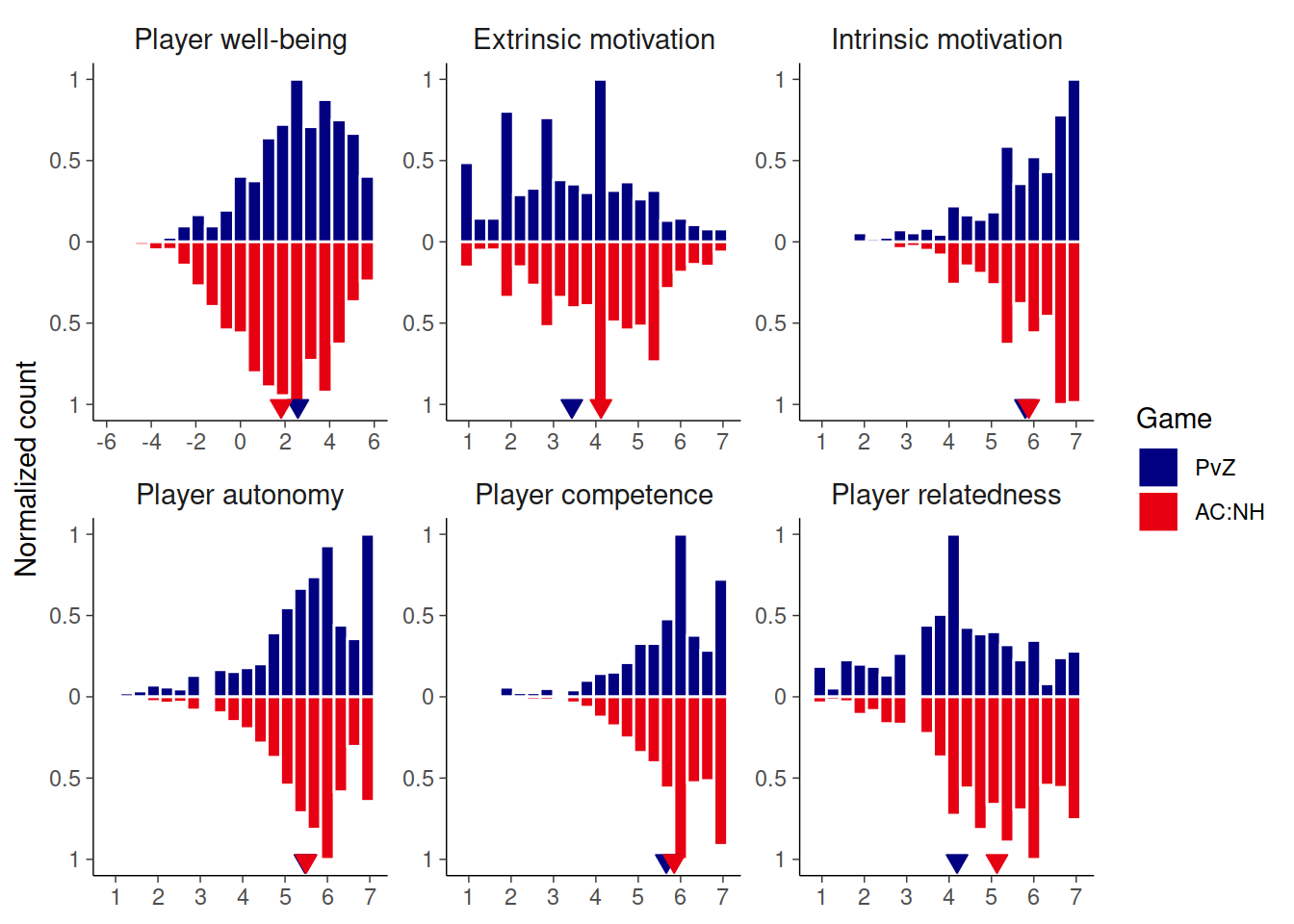
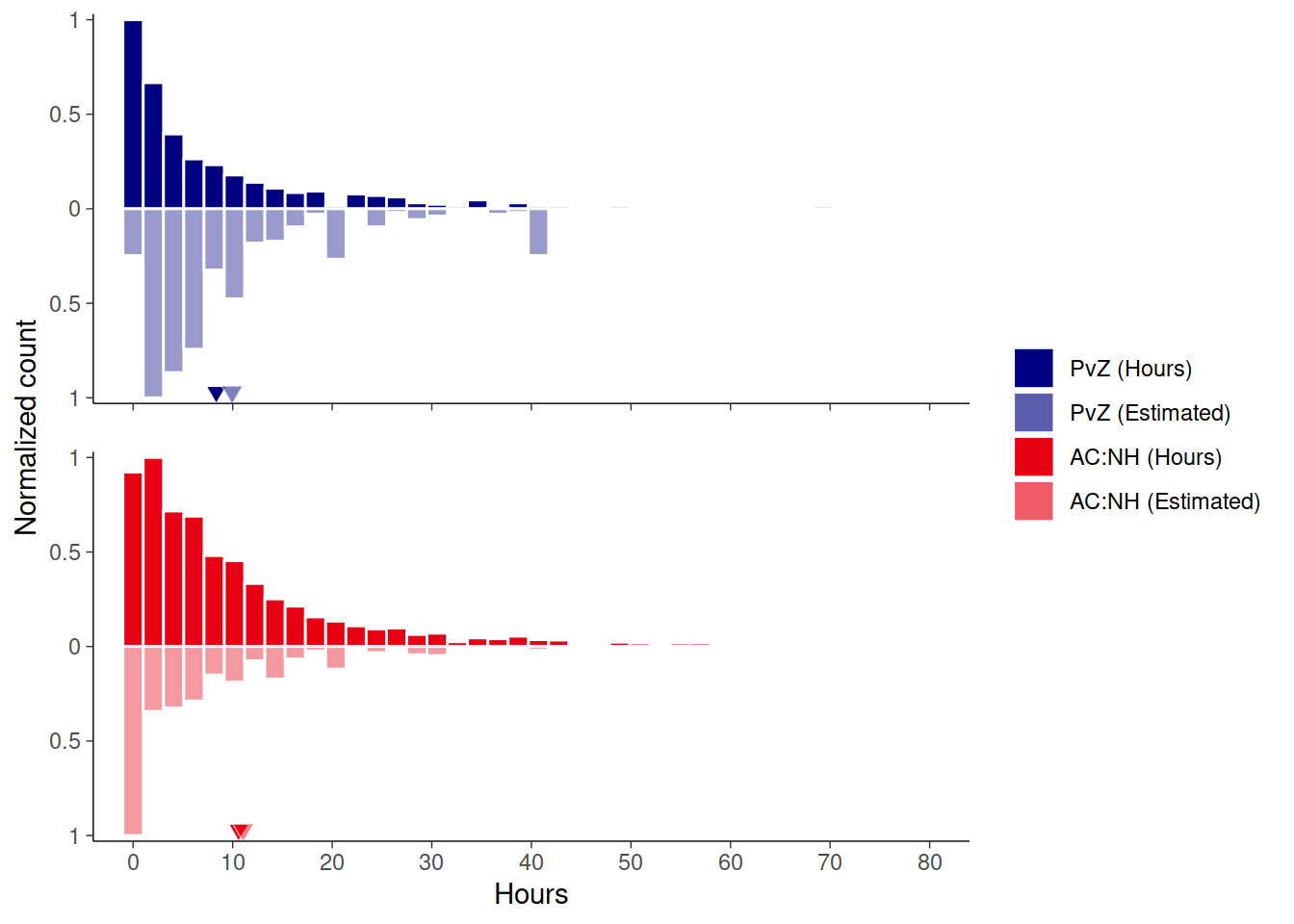
And then a summary figure of the times, subjective and objective. Means indicate means over participants who had both measures.
# Histograms show all participants
tmp <- dat %>%
select(Game, Hours, active_play) %>%
pivot_longer(-Game) %>%
mutate(name = ifelse(name=="Hours", "Hours", "Estimated")) %>%
mutate(Game2 = str_glue("{Game} ({name})"))
# Summaries over people who had both metrics
tmp2 <- dat %>%
select(Game, Hours, active_play) %>%
drop_na(active_play, Hours) %>%
pivot_longer(-Game) %>%
mutate(name = ifelse(name=="Hours", "Hours", "Estimated")) %>%
mutate(Game2 = str_glue("{Game} ({name})")) %>%
group_by(Game, Game2, name) %>%
summarise(m = mean(value, na.rm = TRUE), n = n())
# This is really complicated because of the legend
tmp %>%
ggplot(aes(fill = Game2, alpha = Game2)) +
# Use two variables to control facets and colors/alpha
# Such that legend is constructed appropriately
scale_fill_manual(
values = rep(rev(colors), each = 2),
guide = guide_legend(reverse = TRUE)
) +
scale_alpha_manual(
values = c(.4, 1, .4, 1),
guide = guide_legend(reverse = TRUE)
) +
scale_y_continuous(
breaks = c(-1, -.5, 0, .5, 1), labels = c(1, .5, 0, .5, 1),
expand = expansion(.015)
) +
scale_x_continuous(breaks = pretty_breaks(7)) +
coord_cartesian(ylim = c(-1, 1), xlim = c(0, 80)) +
labs(x = "Hours", y = "Normalized count") +
# Use different geoms to pick appropriate variables
geom_histogram(
data = filter(tmp, name=="Hours"),
aes(x = value, y = stat(ncount)),
bins = 80, col = "white"
) +
geom_histogram(
data = filter(tmp, name=="Estimated"),
aes(x = value, y = stat(ncount) * -1),
bins = 80, col = "white"
) +
geom_point(
data = filter(tmp2, name=="Hours"), aes(x = m, y = -0.97),
shape = 25, size = 2.5, show.legend = FALSE,
col = "white"
) +
geom_point(
data = filter(tmp2, name=="Estimated"), aes(x = m, y = -0.97),
shape = 25, size = 2.5, alpha = 0.5, show.legend = FALSE,
col = "white"
) +
facet_rep_wrap("Game", scales = "fixed", nrow = 2) +
theme(
strip.text = element_blank(),
legend.position = "right",
legend.title = element_blank()
)
# Summary of values truncated from graph
dat %>%
group_by(Game) %>%
summarise(
across(
active_play:Hours,
list(
n = ~sum(!is.na(.x)),
x = ~sum(.x>80, na.rm = TRUE),
p = ~percent(sum(.x>80, na.rm = TRUE) / sum(!is.na(.x)), .01)
)
)
)
#> # A tibble: 2 x 7
#> Game active_play_n active_play_x active_play_p Hours_n Hours_x Hours_p
#> <fct> <int> <int> <chr> <int> <int> <chr>
#> 1 PvZ 517 0 0.00% 469 0 0.00%
#> 2 AC:NH 5927 58 0.98% 2737 7 0.26%
dat %>%
group_by(Game) %>%
summarise(
across(
active_play:Hours,
~max(.x, na.rm = T)
)
)
#> # A tibble: 2 x 3
#> Game active_play Hours
#> <fct> <dbl> <dbl>
#> 1 PvZ 41.0 69.3
#> 2 AC:NH 161. 99.84.6 Correlation matrices
cormat <- function(data, title) {
data %>%
rename_all(str_to_title) %>%
psych::cor.plot(
scale = FALSE, stars = TRUE,
xlas = 2, show.legend = FALSE,
main = str_glue("{title} correlation matrix"),
)
}
select(pvz, where(is.numeric)) %>%
cormat("PvZ")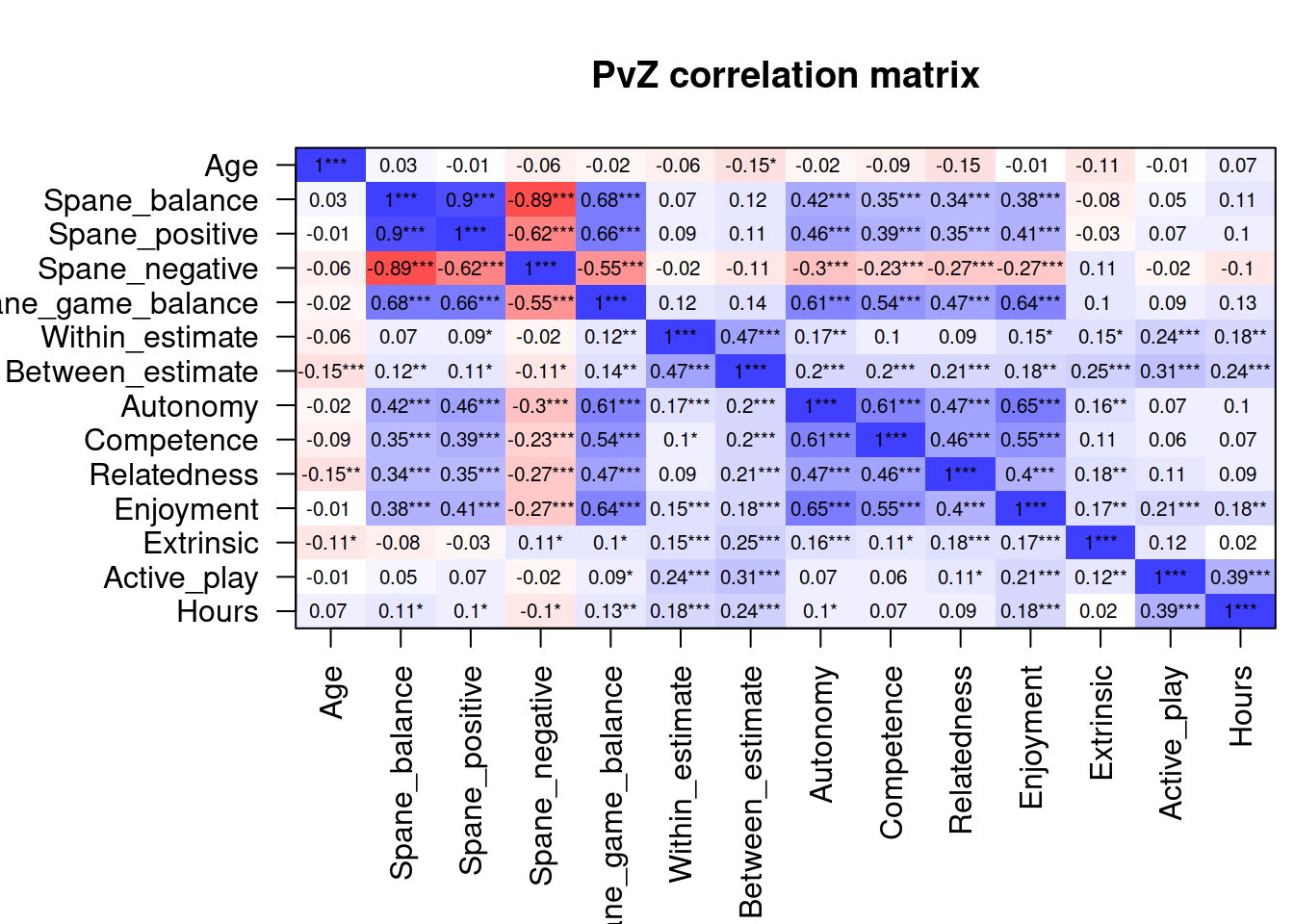
select(ac, where(is.numeric)) %>%
cormat("AC:NH")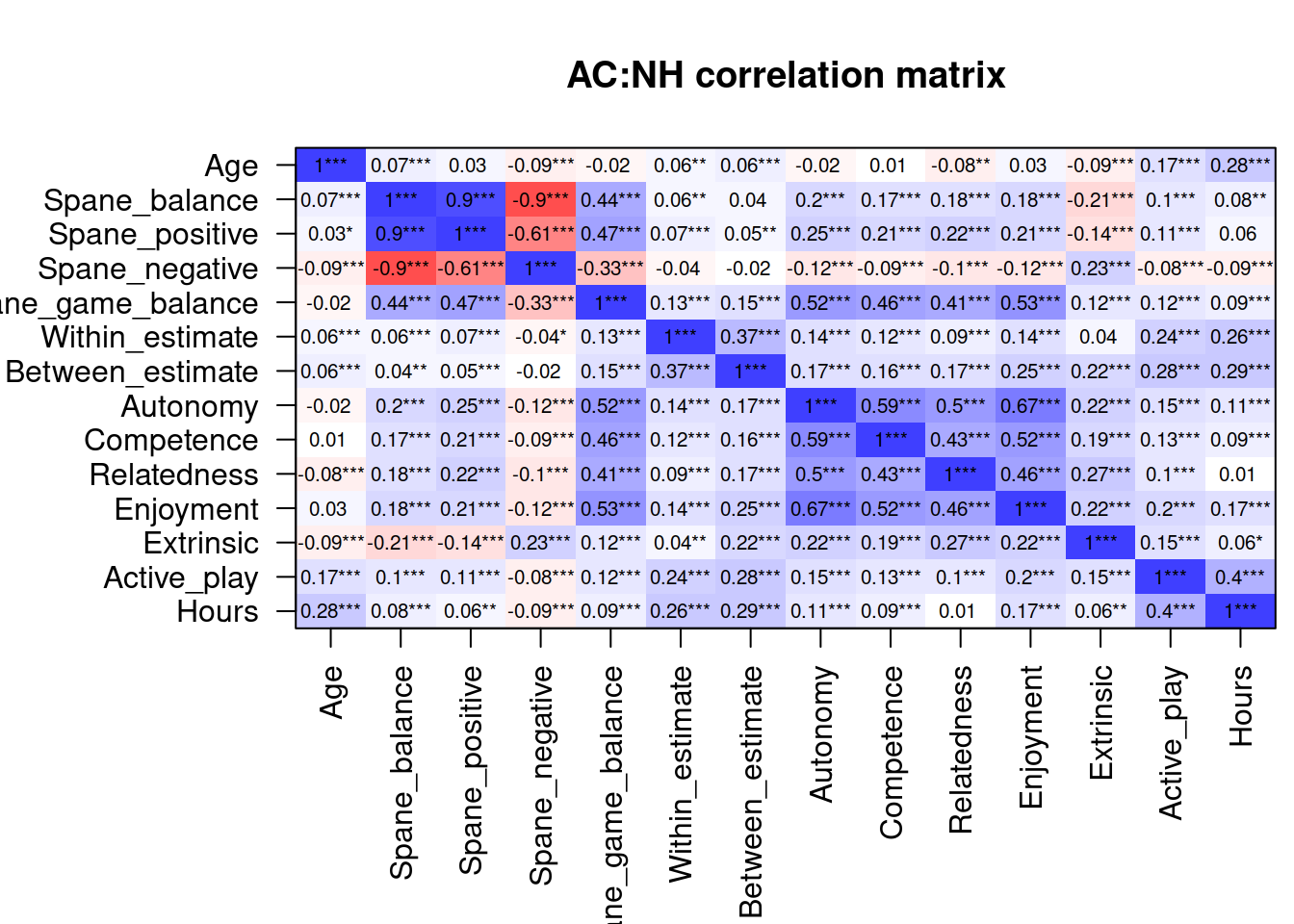
Then we will draw scatterplot matrices, with this custom function:
lm_function <-
function(
data,
mapping,
...
){
p <-
ggplot(
data = data,
mapping = mapping
) +
geom_point(
color = "#56B4E9",
alpha = 0.5
) +
geom_smooth(
method=lm,
fill="#0072B2",
color="#0072B2",
...)
p
}
dens_function <-
function(
data,
mapping,
...
){
p <-
ggplot(
data = data,
mapping = mapping
) +
geom_density(fill = "#009E73", color = NA, alpha = 0.5)
}
pair_plot <-
function(
dat
) {
GGally::ggpairs(
data = dat,
lower = list(continuous = lm_function), # custom helper function
diag = list(continuous = dens_function), # custom helper function
) +
theme(
axis.line=element_blank(),
axis.text.x=element_blank(),
axis.text.y=element_blank(),
axis.ticks=element_blank(),
axis.title.y=element_blank(),
axis.title.x=element_blank(),
legend.position="none",
panel.background=element_blank(),
panel.border=element_blank(),
panel.grid.major=element_blank(),
panel.grid.minor=element_blank(),
plot.background=element_blank(),
strip.background = element_blank()
)
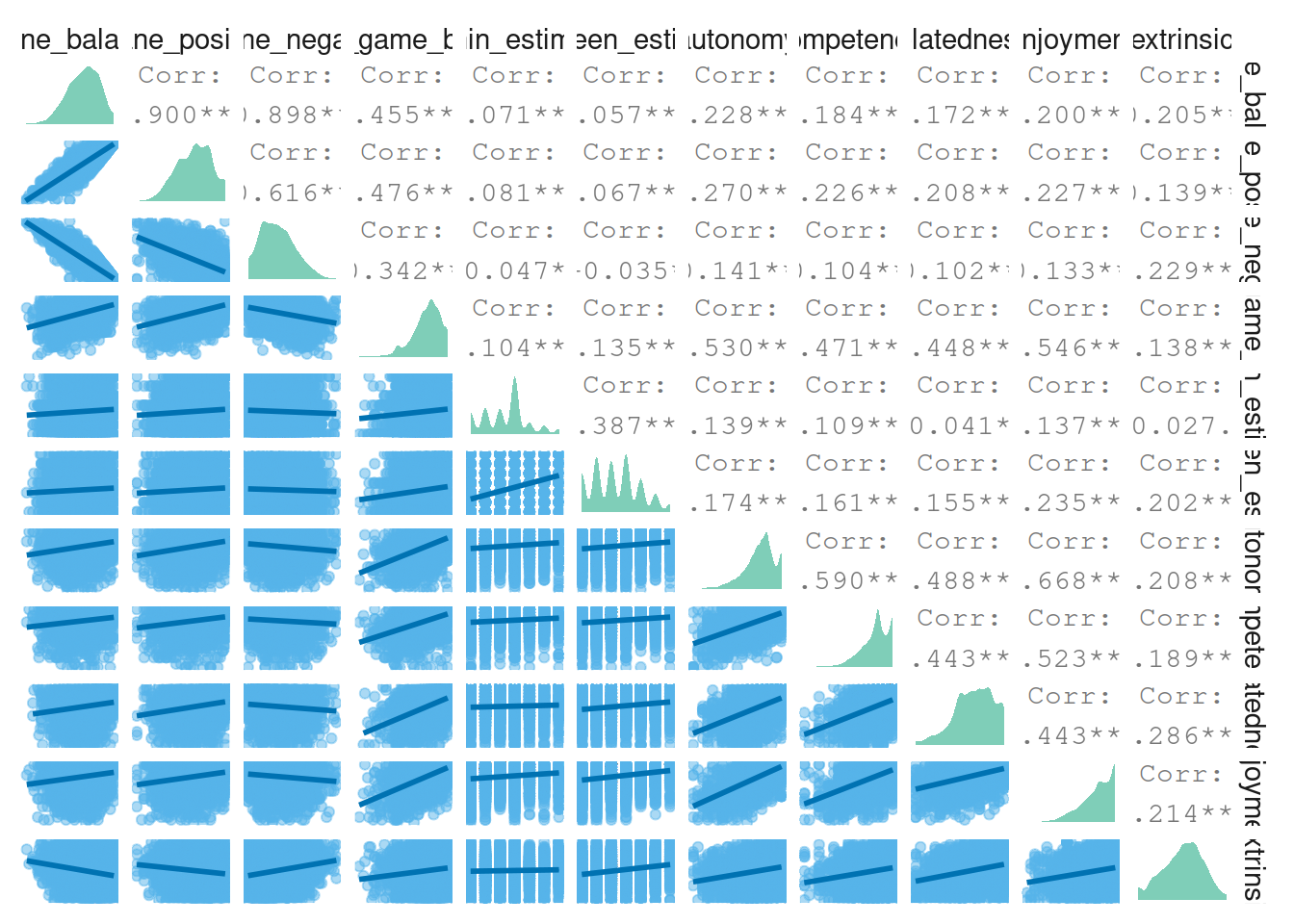
}4.6.1 Well-being & motivations correlations
Let’s have a look at the correlations between the concepts of well-being and motivations (aggregated).
# call the function
pair_plot(
dat%>%
select(
spane_balance:extrinsic,
spane_game_balance
)
)
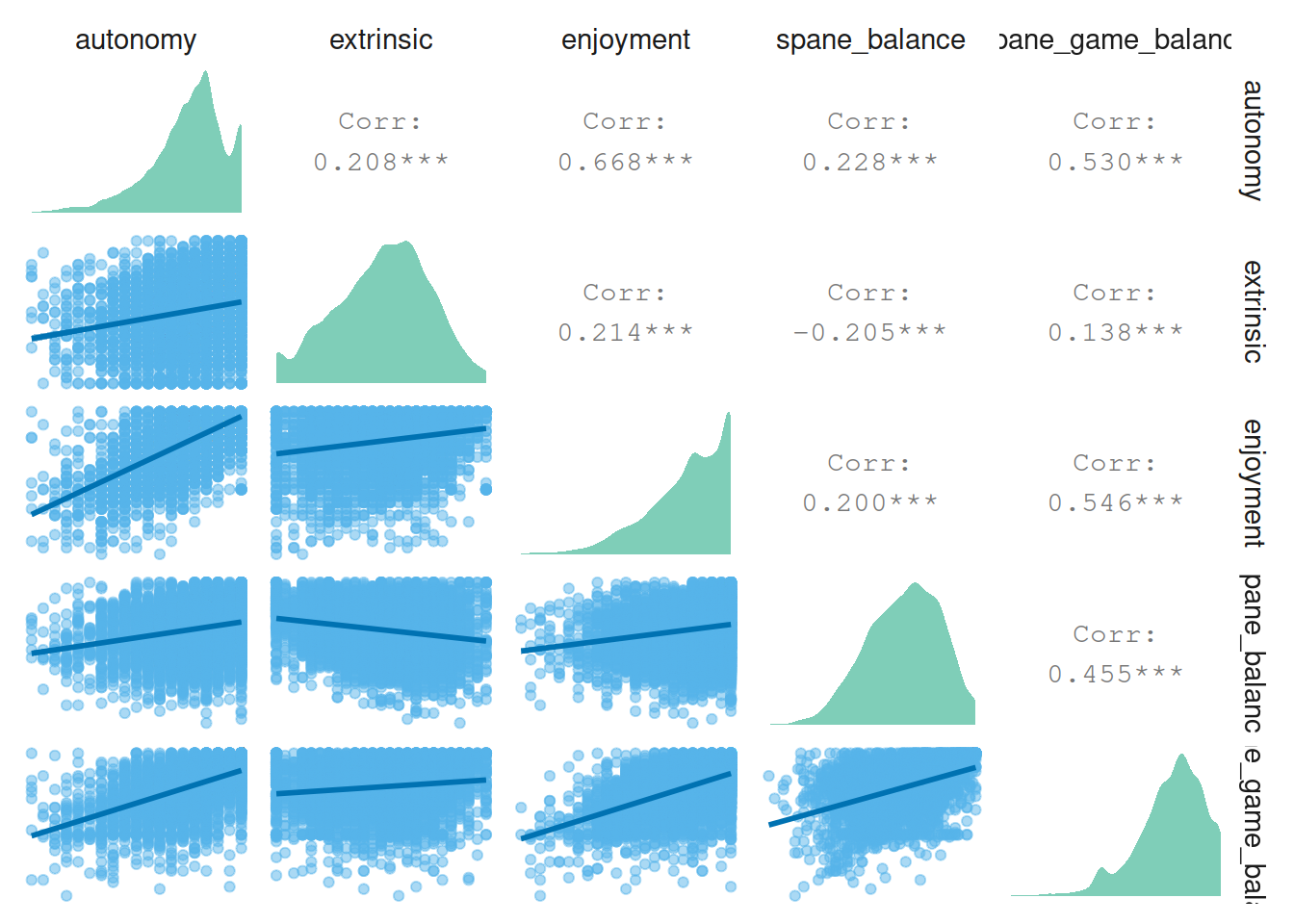
4.6.2 More well-being correlations
Then correlations between well-being, autonomy motivation, enjoyment, and extrinsic motivation.
pair_plot(
dat %>%
select(
autonomy,
extrinsic,
enjoyment,
spane_balance,
spane_game_balance
)
)
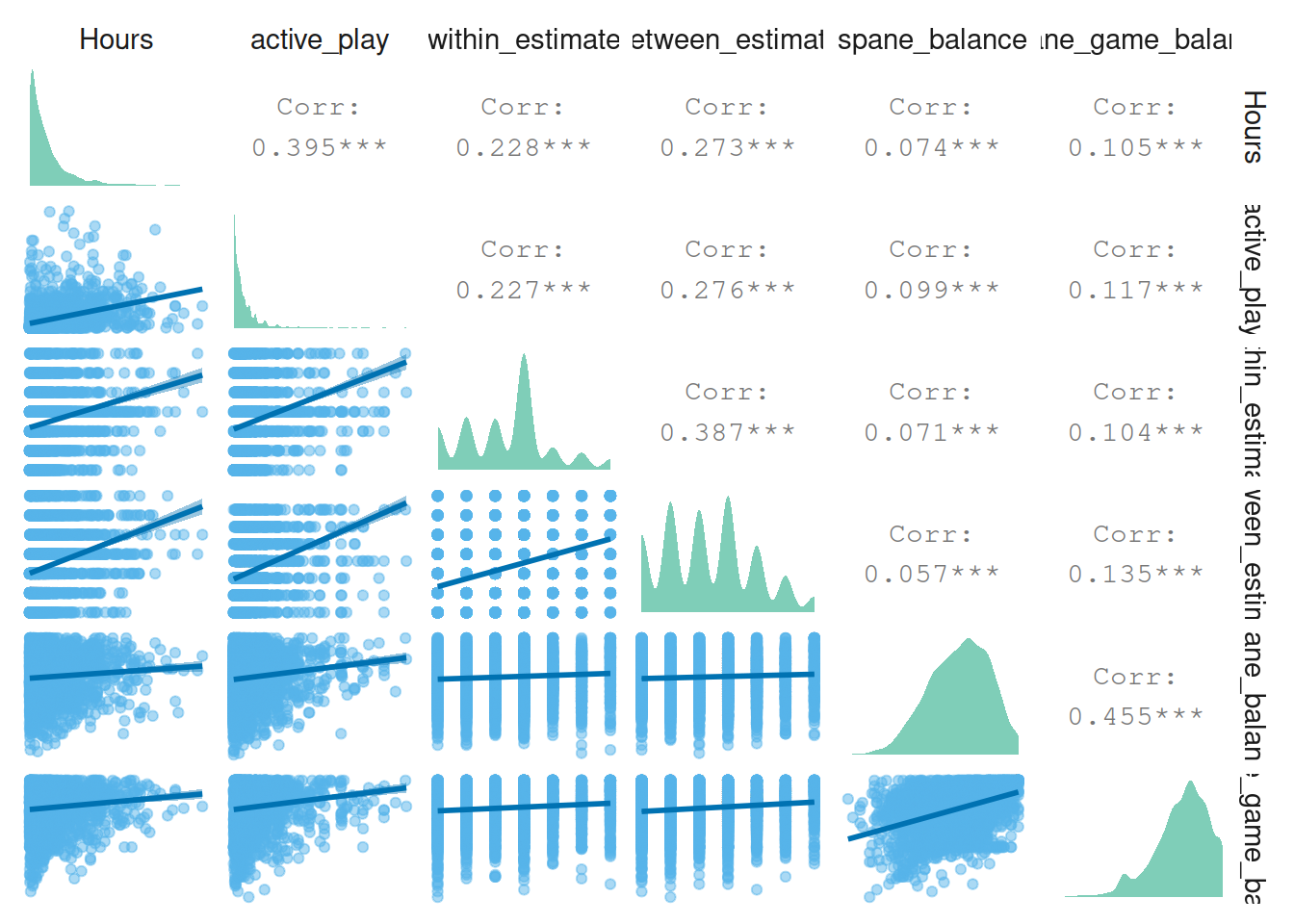
Next relations between self-estimated play (effects) and well-being.
pair_plot(
dat %>%
select(
Hours,
active_play,
within_estimate,
between_estimate,
spane_balance,
spane_game_balance
)
)
4.7 Plot studies combined
Then, let’s plot and describe demographic information. The plots in this section will be for both studies combined.
# raincloud plot function from https://github.com/RainCloudPlots/RainCloudPlots/blob/master/tutorial_R/R_rainclouds.R
# Defining the geom_flat_violin function ----
# Note: the below code modifies the
# existing github page by removing a parenthesis in line 50
"%||%" <- function(a, b) {
if (!is.null(a)) a else b
}
geom_flat_violin <- function(mapping = NULL, data = NULL, stat = "ydensity",
position = "dodge", trim = TRUE, scale = "area",
show.legend = NA, inherit.aes = TRUE, ...) {
layer(
data = data,
mapping = mapping,
stat = stat,
geom = GeomFlatViolin,
position = position,
show.legend = show.legend,
inherit.aes = inherit.aes,
params = list(
trim = trim,
scale = scale,
...
)
)
}
#' @rdname ggplot2-ggproto
#' @format NULL
#' @usage NULL
#' @export
GeomFlatViolin <-
ggproto("GeomFlatViolin", Geom,
setup_data = function(data, params) {
data$width <- data$width %||%
params$width %||% (resolution(data$x, FALSE) * 0.9)
# ymin, ymax, xmin, and xmax define the bounding rectangle for each group
data %>%
group_by(group) %>%
mutate(
ymin = min(y),
ymax = max(y),
xmin = x,
xmax = x + width / 2
)
},
draw_group = function(data, panel_scales, coord) {
# Find the points for the line to go all the way around
data <- transform(data,
xminv = x,
xmaxv = x + violinwidth * (xmax - x)
)
# Make sure it's sorted properly to draw the outline
newdata <- rbind(
plyr::arrange(transform(data, x = xminv), y),
plyr::arrange(transform(data, x = xmaxv), -y)
)
# Close the polygon: set first and last point the same
# Needed for coord_polar and such
newdata <- rbind(newdata, newdata[1, ])
ggplot2:::ggname("geom_flat_violin", GeomPolygon$draw_panel(newdata, panel_scales, coord))
},
draw_key = draw_key_polygon,
default_aes = aes(
weight = 1, colour = "grey20", fill = "white", size = 0.5,
alpha = NA, linetype = "solid"
),
required_aes = c("x", "y")
)
# function that returns summary stats
describe <- function(
dat,
variable,
trait = FALSE
){
# if variable is not repeated-measures, take only one measure per participant
if (trait == TRUE){
dat <-
dat %>%
group_by(player_id) %>%
slice(1) %>%
ungroup()
}
# then get descriptives
descriptives <-
dat %>%
filter(!is.na(UQ(sym(variable)))) %>% # remove missing values
summarise(
across(
!! variable,
list(
N = ~ n(),
mean = mean,
sd = sd,
median = median,
min = min,
max = max,
cilow = ~Rmisc::CI(.x)[[3]], # lower CI
cihigh = ~Rmisc::CI(.x)[[1]] # upper CI
)
)
)
descriptives <-
descriptives %>%
# only keep measure
rename_all(
~ str_remove(
.,
paste0(variable, "_")
)
) %>%
mutate(
variable = variable,
range = max - min
) %>%
relocate(variable) %>%
relocate(
range,
.after = max
)
return(descriptives)
}
single_cloud <-
function(
raw_data,
summary_data,
variable,
color,
title,
trait = FALSE
){
# take only one row per person if it's a trait variable
if (trait == TRUE){
raw_data <-
raw_data %>%
group_by(player_id) %>%
slice(1) %>%
ungroup()
}
# the plot
p <-
ggplot(
raw_data %>%
mutate(Density = 1),
aes(
x = Density,
y = get(variable)
)
) +
geom_flat_violin( # the "cloud"
position = position_nudge(x = .2, y = 0),
adjust = 2,
color = NA,
fill = color,
alpha = 0.5
) +
geom_point( # the "rain"
position = position_jitter(width = .15),
size = 1,
color = color,
alpha = 0.5
) +
geom_point( # the mean from the summary stats
data = summary_data %>%
filter(variable == !! variable) %>%
mutate(Density = 1),
aes(
x = Density + 0.175,
y = mean
),
color = color,
size = 2.5
) +
geom_errorbar( # error bars
data = summary_data %>%
filter(variable == !! variable) %>%
mutate(Density = 1),
aes(
x = Density + 0.175,
y = mean,
ymin = cilow,
ymax = cihigh
),
width = 0,
size = 0.8,
color = color
) +
ylab(title) +
theme_cowplot() +
theme(
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.ticks.x = element_blank(),
axis.title.y = element_blank(),
axis.line = element_blank()
) +
guides(
color = FALSE,
fill = FALSE
) +
coord_flip()
return(p)
}
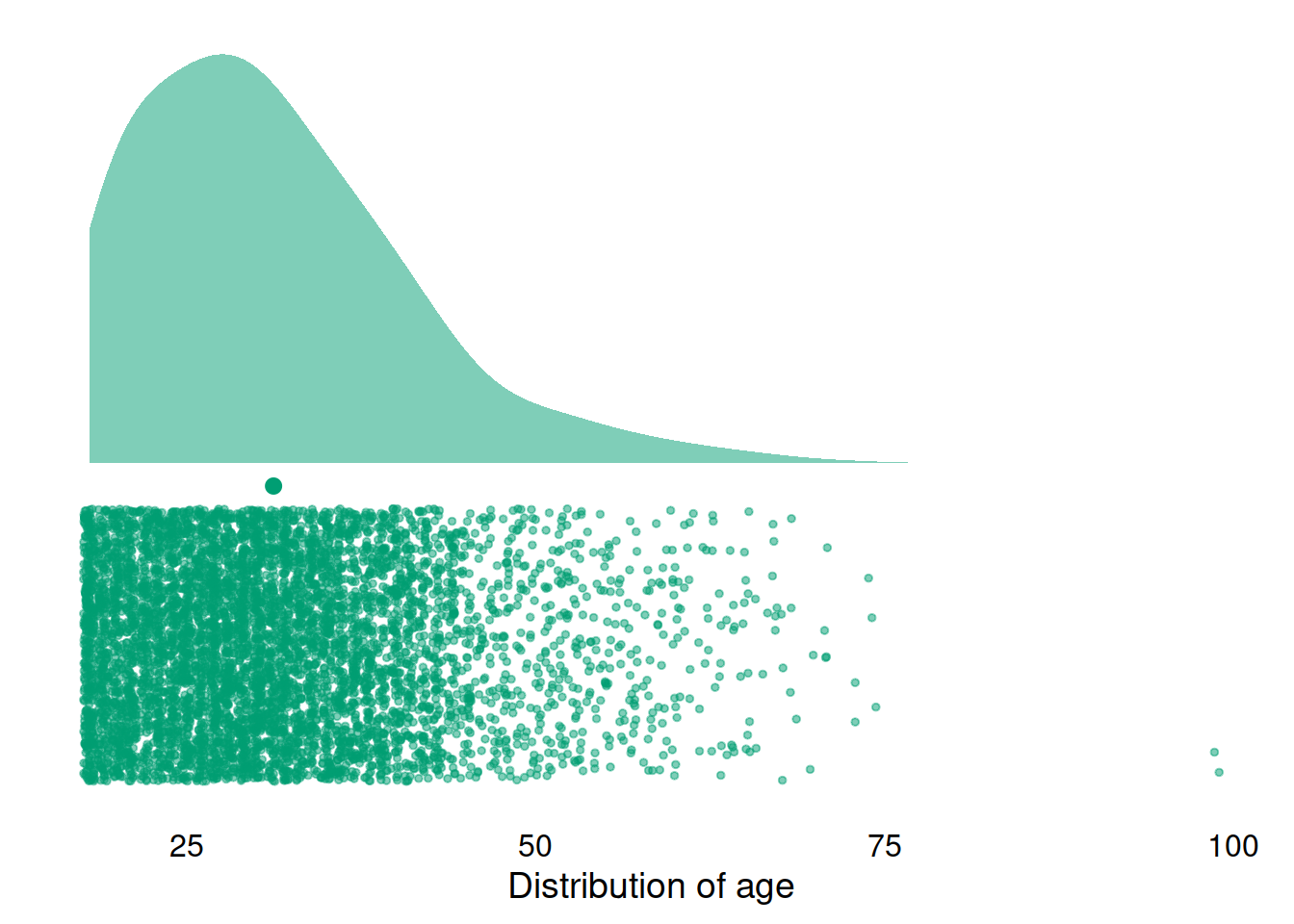
single_cloud(
dat,
describe(dat, "age", trait = FALSE),
"age",
"#009E73",
"Distribution of age"
)
And a check of the variable distributions.
p1 <-
single_cloud(
pvz,
describe(pvz, "spane_balance", trait = TRUE),
"spane_balance",
"#56B4E9",
title = "SPANE Balance",
trait = TRUE
)
p2 <-
single_cloud(
pvz,
describe(pvz, "autonomy", trait = TRUE),
"autonomy",
"#009E73",
title = "Autonomy",
trait = TRUE
)
p3 <-
single_cloud(
pvz,
describe(pvz, "competence", trait = TRUE),
"competence",
"#F0E442",
title = "Competence",
trait = TRUE
)
p4 <-
single_cloud(
pvz,
describe(pvz, "relatedness", trait = TRUE),
"relatedness",
"#000000",
title = "Relatedness",
trait = TRUE
)
p5 <-
single_cloud(
pvz,
describe(pvz, "enjoyment", trait = TRUE),
"enjoyment",
"#0072B2",
title = "Enjoyment",
trait = TRUE
)
p6 <-
single_cloud(
pvz,
describe(pvz, "extrinsic", trait = TRUE),
"extrinsic",
"#D55E00",
title = "Extrinsic motivation",
trait = TRUE
)
p7 <-
single_cloud(
pvz,
describe(pvz, "active_play", trait = TRUE),
"active_play",
"#CC79A7",
title = "Self-estimated play",
trait = TRUE
)
(p1 | p2) / (p3 | p4) / (p5 | p6) / p7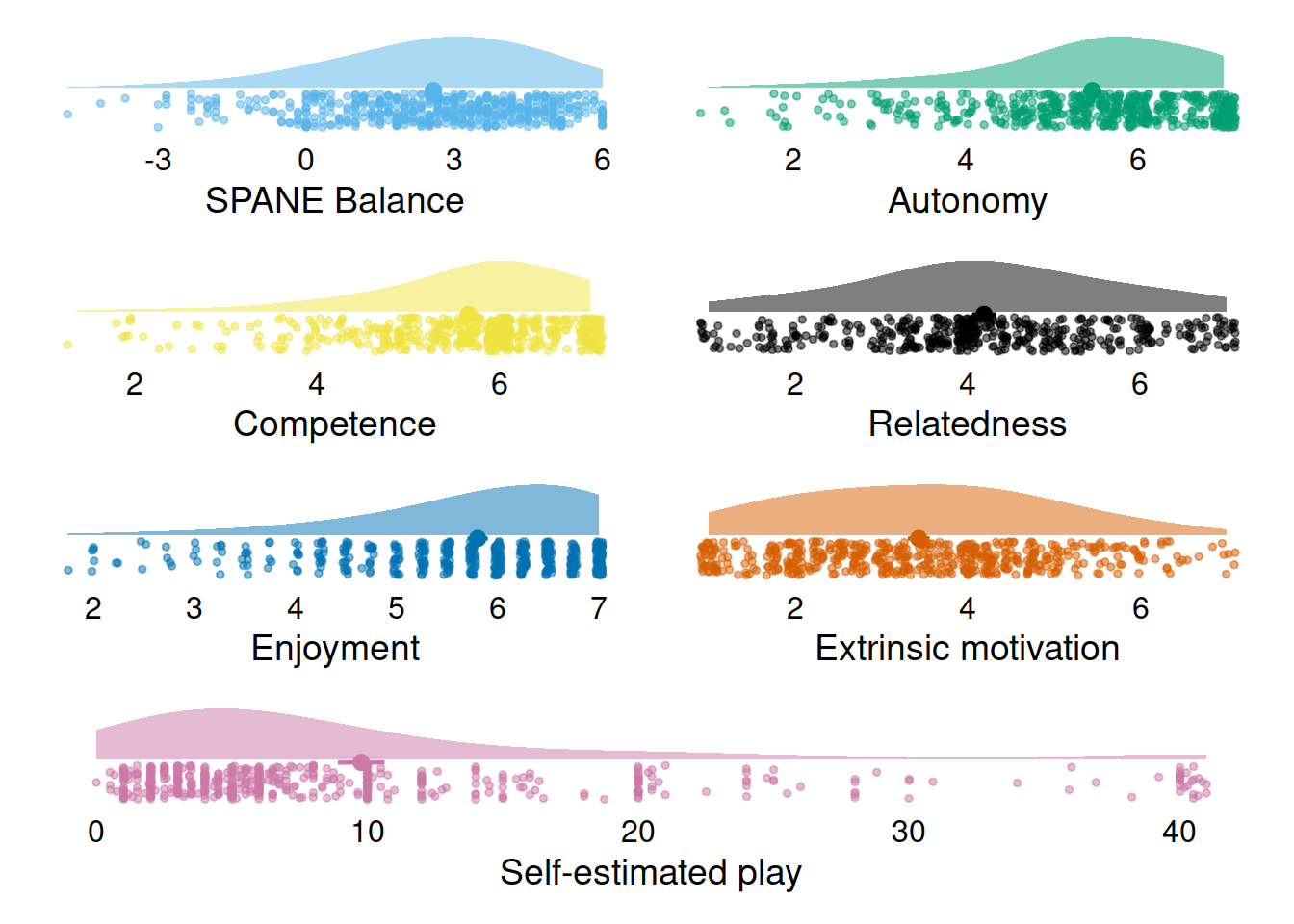
4.8 Create summary figure
Alright, here I’ll create a summary figure that shows a beeswarm plot for each of the variables we’re interested in.
First, because we’ll plot per measure, let’s turn the data into the long format.
dat_long <-
dat %>%
pivot_longer(
spane_balance:Hours,
names_to = "Variable",
values_to = "value"
) %>%
mutate(Variable = as.factor(Variable))Now, let’s get summary stats for each measure.
# function to get summary stats
get_summary <-
function(
dat
) {
summary_stats <-
dat %>%
group_by(Game) %>%
summarise(
across(
spane_balance:Hours,
list(
N = ~ sum(!is.na(.x)),
Mean = ~ mean(.x, na.rm = TRUE),
SD = ~ sd(.x, na.rm = TRUE),
Median = ~ median(.x, na.rm = TRUE),
Min = ~ min(.x, na.rm = TRUE),
Max = ~ max(.x, na.rm = TRUE),
`Lower 95%CI` = ~Rmisc::CI(na.omit(.x))[[3]], # lower CI
`Upper 95%CI` = ~Rmisc::CI(na.omit(.x))[[1]]
)
)
) %>%
# get into long format
pivot_longer(
-Game,
names_to = c("Variable", "Measure"),
values_to = "Value",
names_pattern = "(.*)_([^_]+$)" # match by last occurrence of underscore
) %>%
# and back to wide format
pivot_wider(
id_cols = c(Game, Variable),
names_from = Measure,
values_from = Value
)
return(summary_stats)
}
summary_stats <- get_summary(dat)
# rename both so that we have the same order of factor levels for the figure
rename_and_relevel <-
function(
dat
) {
dat <-
dat %>%
mutate(
Variable = fct_relevel(
Variable,
"SPANE" = "spane_balance",
"Hours",
"active_play",
"autonomy",
"competence",
"relatedness",
"enjoyment",
"extrinsic"
),
Variable = fct_recode(
Variable,
"Affective well-being" = "spane_balance",
"Objective play time" = "Hours",
"Self-reported play time" = "active_play",
"Autonomy" = "autonomy",
"Competence" = "competence",
"Relatedness" = "relatedness",
"Enjoyment" = "enjoyment",
"Extrinsic" = "extrinsic"
)
)
return(dat)
}
dat_long <- rename_and_relevel(dat_long)
summary_stats <- rename_and_relevel(summary_stats)And then the beeswarm plots.
summary_stats <-
summary_stats %>%
group_by(Variable) %>%
mutate( # the position of geom_text
x = max(Max) * 0.85,
across(
c(Mean, SD),
~ format(round(.x, digits = 1), nsmall = 1)
)
)
# a function because we "split" up the plot with patchwork
beeswarm_plots <-
function(
dat,
subset,
mean_position,
sd_position,
col_font_size,
row_font_size
){
ggplot(
dat %>%
filter(
Variable %in% subset
),
aes(
x = value,
y = 1,
color = Game
)
) +
geom_quasirandom(
size = 1,
groupOnX = FALSE,
shape = 1,
alpha = 0.8
) +
facet_grid(
Game ~ Variable,
scales = "free_x"
) +
geom_text(
data = summary_stats %>%
filter(Variable %in% subset),
aes(
x = x,
y = mean_position,
label = paste0("M = ", Mean)
),
color = "black",
size = 2.5
) +
geom_text(
data = summary_stats %>%
filter(Variable %in% subset),
aes(
x = x,
y = sd_position,
label = paste0("SD = ", SD)
),
color = "black",
size = 2.5
) +
facet_rep_grid(
Game ~ Variable,
scales = "free_x"
) +
scale_color_manual(values=c("navyblue", "#E60012")) +
theme(
axis.text.y = element_blank(),
axis.title.x = element_blank(),
axis.ticks.y = element_blank(),
axis.title.y = element_blank(),
axis.line = element_line(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
strip.background.x = element_blank(),
strip.background.y = element_blank(),
strip.text.x = element_text(size = col_font_size),
strip.text.y = element_text(size = row_font_size),
legend.position = "none"
) -> p
return(p)
}
p1 <-
beeswarm_plots(
dat_long,
c("Competence", "Relatedness", "Enjoyment", "Extrinsic", "Autonomy"),
1.65,
1.5,
10,
11
)
p2 <-
beeswarm_plots(
dat_long,
c("Affective well-being", "Objective play time", "Self-reported play time"),
2.0,
1.8,
10,
11
)
p2 / p1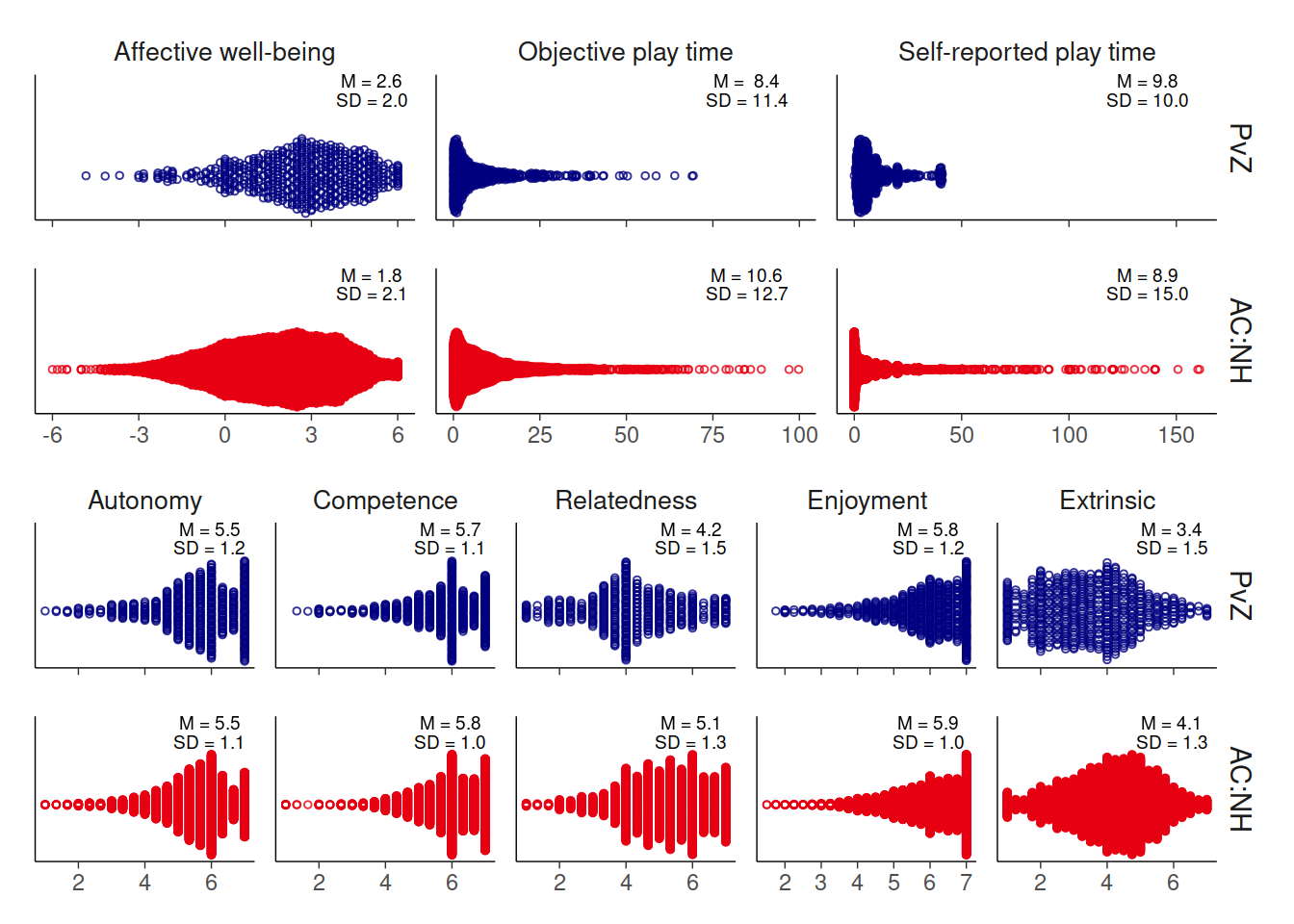
4.9 Session info
sessionInfo()
#> R version 4.0.3 (2020-10-10)
#> Platform: x86_64-pc-linux-gnu (64-bit)
#> Running under: Ubuntu 20.04.1 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/liblapack.so.3
#>
#> locale:
#> [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
#> [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
#> [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
#> [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] forcats_0.5.0 stringr_1.4.0 dplyr_1.0.2 purrr_0.3.4
#> [5] readr_1.4.0 tidyr_1.1.2 tibble_3.0.4 tidyverse_1.3.0
#> [9] lubridate_1.7.9.2 lemon_0.4.5 ggbeeswarm_0.6.0 ggplot2_3.3.2
#> [13] janitor_2.0.1 cowplot_1.1.0 sjPlot_2.8.6 patchwork_1.1.0
#> [17] scales_1.1.1 visdat_0.5.3 here_1.0.1 knitr_1.30
#> [21] pacman_0.5.1
#>
#> loaded via a namespace (and not attached):
#> [1] nlme_3.1-150 fs_1.5.0 insight_0.11.1 httr_1.4.2
#> [5] rprojroot_2.0.2 tools_4.0.3 backports_1.2.1 utf8_1.1.4
#> [9] R6_2.5.0 sjlabelled_1.1.7 vipor_0.4.5 DBI_1.1.0
#> [13] colorspace_2.0-0 withr_2.3.0 mnormt_2.0.2 tidyselect_1.1.0
#> [17] gridExtra_2.3 emmeans_1.5.3 compiler_4.0.3 cli_2.2.0
#> [21] performance_0.6.1 rvest_0.3.6 xml2_1.3.2 sandwich_3.0-0
#> [25] labeling_0.4.2 bookdown_0.21 bayestestR_0.8.0 psych_2.0.9
#> [29] mvtnorm_1.1-1 digest_0.6.27 minqa_1.2.4 rmarkdown_2.6
#> [33] pkgconfig_2.0.3 htmltools_0.5.0 lme4_1.1-26 highr_0.8
#> [37] dbplyr_2.0.0 Rmisc_1.5 rlang_0.4.9 readxl_1.3.1
#> [41] rstudioapi_0.13 farver_2.0.3 generics_0.1.0 zoo_1.8-8
#> [45] jsonlite_1.7.2 magrittr_2.0.1 parameters_0.10.1 Matrix_1.2-18
#> [49] fansi_0.4.1 Rcpp_1.0.5 munsell_0.5.0 lifecycle_0.2.0
#> [53] stringi_1.5.3 multcomp_1.4-15 yaml_2.2.1 snakecase_0.11.0
#> [57] MASS_7.3-53 plyr_1.8.6 grid_4.0.3 parallel_4.0.3
#> [61] sjmisc_2.8.5 crayon_1.3.4 lattice_0.20-41 ggeffects_1.0.1
#> [65] haven_2.3.1 splines_4.0.3 sjstats_0.18.0 hms_0.5.3
#> [69] tmvnsim_1.0-2 pillar_1.4.7 boot_1.3-25 estimability_1.3
#> [73] effectsize_0.4.1 codetools_0.2-18 reprex_0.3.0 glue_1.4.2
#> [77] evaluate_0.14 modelr_0.1.8 vctrs_0.3.5 nloptr_1.2.2.2
#> [81] cellranger_1.1.0 gtable_0.3.0 assertthat_0.2.1 xfun_0.19
#> [85] xtable_1.8-4 broom_0.7.2 coda_0.19-4 survival_3.2-7
#> [89] beeswarm_0.2.3 statmod_1.4.35 TH.data_1.0-10 ellipsis_0.3.1