4 Main analysis
This document contains the main analyses presented in the manuscript.
We first load the required R packages.
library(knitr)
library(scales)
library(DT)
library(kableExtra)
library(ggpp)
library(bayestestR)
library(brms)
library(ragg)
library(here)
library(ggtext)
library(ggstance)
library(ggdist)
library(memoise)
library(cachem)
library(patchwork)
library(lavaan)
library(multidplyr)
library(tidyverse)
library(tidybayes)
library(janitor)
cd <- cache_disk("memoise")Define plotting, table, and parallel processing options.
# parallel computations
MAX_CORES <- as.numeric(Sys.getenv("MAX_CORES"))
if (is.na(MAX_CORES)) MAX_CORES <- parallel::detectCores(logical = FALSE)
cluster <- new_cluster(MAX_CORES)
# load packages on clusters
cluster_library(cluster, c("dplyr", "lavaan"))
# MCMC settings
options(mc.cores = 1)
if (require("cmdstanr")) options(brms.backend = "cmdstanr")
# Save outputs here
dir.create("models", FALSE)
# Functions in R/
source(here("R", "meta-analysis-helpers.R"))We analyse the data cleaned previously.
data_path <- here("Data", "cleaned_data.rds")
if (file.exists(data_path)) {
d <- read_rds(file = data_path)
} else {
stop(str_glue("{data_path} doesn't exist, run `01-clean.Rmd` to create it."))
}
# Make wave a nicely labelled factor
d <- d %>%
mutate(Wave = factor(wid, levels = 1:3, labels = paste0("Wave ", 1:3)))
# Rename game to fit titles in plots
d <- d %>%
mutate(Game = if_else(Game == "Gran Turismo Sport", "GT Sport", Game))For all analyses, game time indicates average number of hours per day. The telemetry indicates total hours in the two week window, so we divide that by 14 to talk about average hours per day.
d <- d %>%
mutate(
Hours = Hours / 14,
hours_est = hours_est / 14
)4.1 Variables over time
Hours played per week is cut at a reasonable value for the figures:
d %>%
mutate(Hours_over_3 = Hours > 3) %>%
tabyl(Hours_over_3) %>%
adorn_pct_formatting() %>%
kbl(caption = "Table of hours excluded from figure") %>%
kable_classic(full_width = FALSE, html_font = Font)| Hours_over_3 | n | percent | valid_percent |
|---|---|---|---|
| FALSE | 113595 | 97.3% | 97.4% |
| TRUE | 3075 | 2.6% | 2.6% |
| NA | 100 | 0.1% |
|
tmp <- d %>%
select(
Game, Wave, pid,
Hours, Affect, `Life satisfaction`,
Intrinsic, Extrinsic
) %>%
pivot_longer(Hours:Extrinsic) %>%
drop_na(value) %>%
filter(!(name == "Hours" & value > 3)) %>%
mutate(name = if_else(name == "Hours", "Hours / day", name)) %>%
mutate(name = fct_inorder(name))
tmp %>%
ggplot(
aes(
Wave,
value
)
) +
scale_x_discrete(labels = 1:3, expand = expansion(c(0.1, .1))) +
scale_y_continuous(
"Value",
breaks = pretty_breaks(),
expand = expansion(.025)
) +
geom_blank() +
stat_halfeye(
height = .02,
normalize = "panels",
slab_color = col1,
slab_fill = alpha(col1, 0.3),
slab_size = 0.5,
adjust = 1.1,
point_interval = NULL,
show.legend = FALSE
) +
stat_summary(
fun.data = mean_cl_normal,
fatten = 1.25
) +
stat_summary(
fun = mean,
geom = "line",
size = .33,
group = 1
) +
facet_grid(name ~ Game, scales = "free_y") +
theme(
legend.position = "none"
)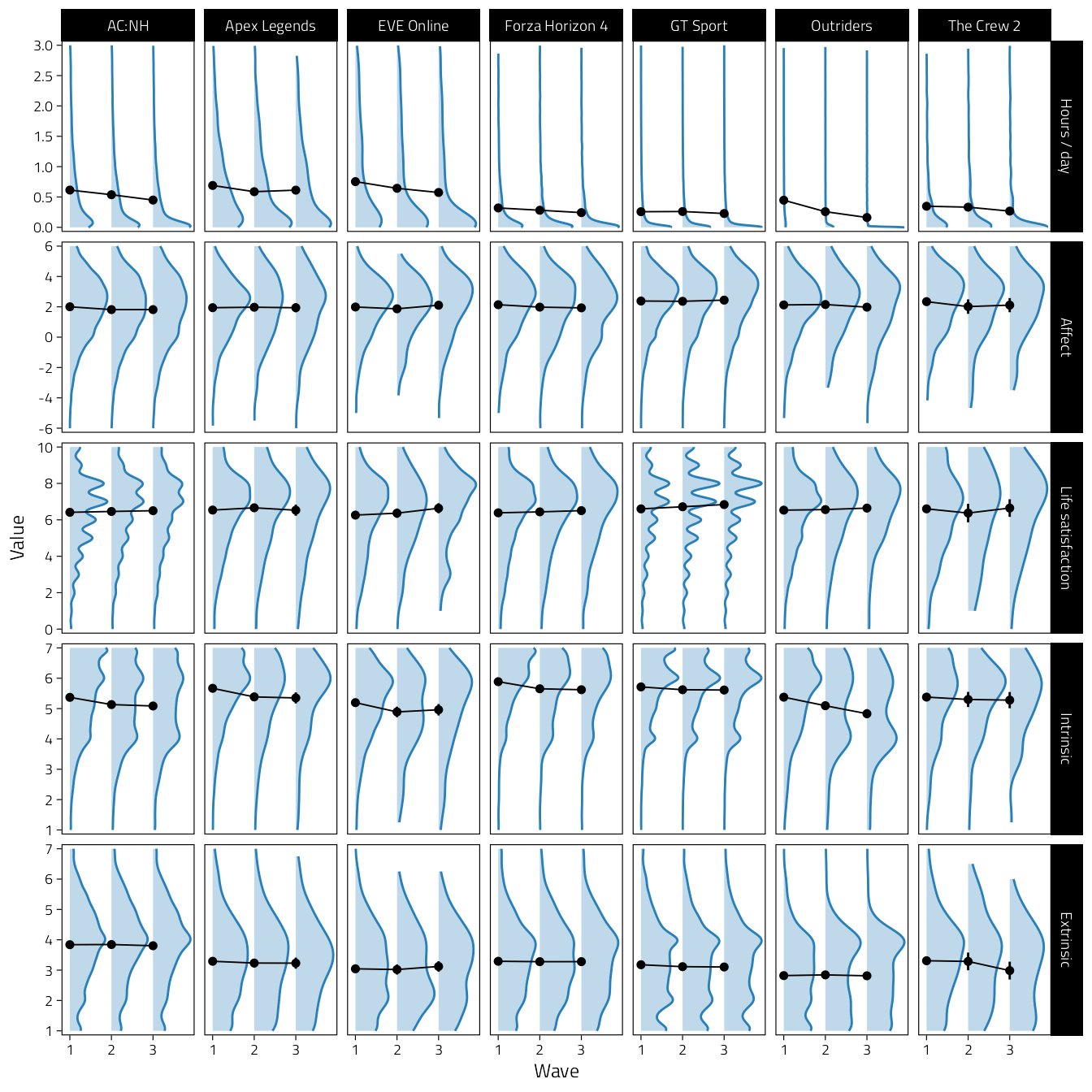
Figure 4.1: Density plots of key variables over time.
Then take a look at a simple model of change over time for each variable. Note that we can’t use varying slopes with lmer because there’s not enough data, so just random intercepts for players.
library(lme4)
library(broom.mixed)
path <- "models/parameters_change_over_time.rds"
if (!file.exists(path)) {
parameters_change_over_time <- d %>%
select(
Game, pid, wid,
Hours, Intrinsic, Extrinsic,
Affect, `Life satisfaction`
) %>%
# Put intercept at first wave
mutate(Wave = wid - 1) %>%
pivot_longer(Hours:`Life satisfaction`, names_to = "Variable") %>%
group_by(Variable) %>%
summarise(
lmer(value ~ Wave + (1 | pid) + (1 + Wave | Game), data = cur_data()) %>%
tidy(., "fixed", conf.int = TRUE)
)
saveRDS(parameters_change_over_time, path)
} else {parameters_change_over_time <- readRDS(path)}
parameters_change_over_time %>%
mutate(across(where(is.numeric), ~ format(round(.x, 2), nsmall = 2))) %>%
mutate(Result = str_glue("{estimate}, 95%CI [{conf.low}, {conf.high}]")) %>%
select(Variable, term, Result) %>%
pivot_wider(names_from = term, values_from = Result) %>%
kbl(caption = "Change over time parameter estimates") %>%
kable_classic(full_width = FALSE, html_font = Font)| Variable | (Intercept) | Wave |
|---|---|---|
| Affect | 2.12, 95%CI [ 2.00, 2.24] | -0.05, 95%CI [-0.10, -0.01] |
| Extrinsic | 3.25, 95%CI [ 3.02, 3.48] | -0.01, 95%CI [-0.03, 0.01] |
| Hours | 0.69, 95%CI [ 0.39, 1.00] | -0.09, 95%CI [-0.15, -0.04] |
| Intrinsic | 5.51, 95%CI [ 5.33, 5.69] | -0.18, 95%CI [-0.23, -0.13] |
| Life satisfaction | 6.47, 95%CI [6.40, 6.55] | 0.05, 95%CI [0.03, 0.08] |
4.2 Simple correlation
Before more informative modelling, we estimate simple bivariate regressions between the key variables.
tmp <- d %>%
arrange(pid, wid) %>%
group_by(pid) %>%
mutate(h1 = lag(Hours))
fit <- lmer(Affect ~ h1 + (1 | pid) + (h1 | Game), data = tmp)
summary(fit)## Linear mixed model fit by REML ['lmerMod']
## Formula: Affect ~ h1 + (1 | pid) + (h1 | Game)
## Data: tmp
##
## REML criterion at convergence: 101413.9
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -4.4954 -0.3566 0.0485 0.3947 3.9020
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## pid (Intercept) 3.391653 1.84164
## Game (Intercept) 0.045830 0.21408
## h1 0.001198 0.03461 -1.00
## Residual 0.992279 0.99613
## Number of obs: 25181, groups: pid, 17197; Game, 7
##
## Fixed effects:
## Estimate Std. Error t value
## (Intercept) 1.97604 0.08972 22.024
## h1 0.07184 0.02063 3.483
##
## Correlation of Fixed Effects:
## (Intr)
## h1 -0.750
## optimizer (nloptwrap) convergence code: 0 (OK)
## boundary (singular) fit: see ?isSingular
d %>%
select(
Game, pid, wid, Wave,
Hours, hours_est, Intrinsic, Extrinsic,
Affect, `Life satisfaction`
) %>%
pivot_longer(
c(Affect, `Life satisfaction`),
names_to = "Outcome", values_to = "Outcome_value"
) %>%
pivot_longer(
c(Hours, hours_est, Intrinsic, Extrinsic),
names_to = "Predictor", values_to = "Predictor_value"
) %>%
group_by(Game, Outcome, Predictor, Wave) %>%
summarise(
tidy(
lm(
Outcome_value ~ Predictor_value,
data = cur_data()
),
conf.int = TRUE
),
) %>%
filter(term != "(Intercept)") %>%
ggplot(aes(estimate, Game, col = Wave)) +
geom_vline(xintercept = 0, lty = 2, size = .25) +
scale_x_continuous(
"Bivariate regression coefficient (95%CI)",
breaks = pretty_breaks()
) +
geom_pointrangeh(
aes(xmin = conf.low, xmax = conf.high),
size = .4,
position = position_dodge2v(.35)
) +
facet_grid(Outcome ~ Predictor) +
theme(
legend.position = "bottom",
axis.title.y = element_blank()
)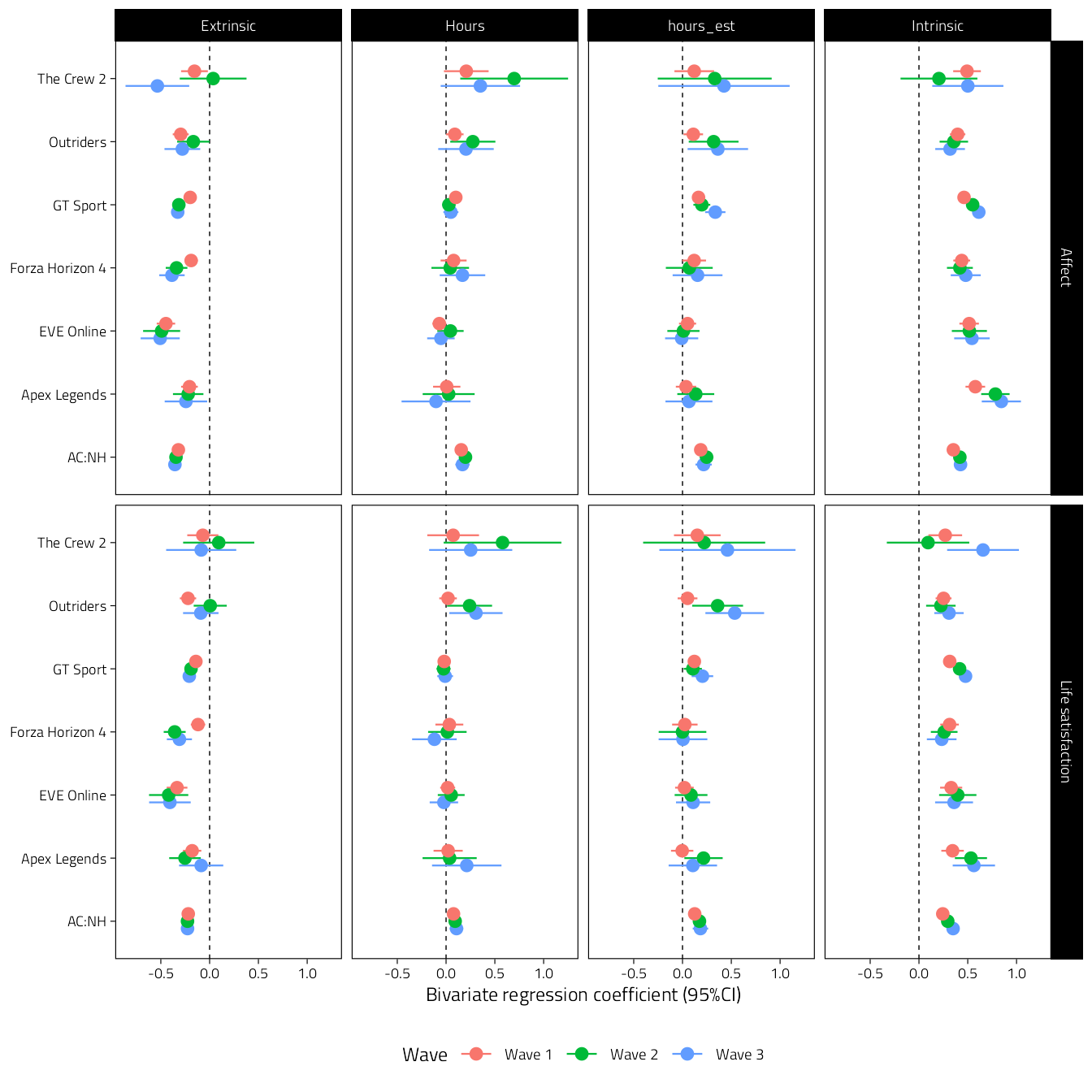
Figure 4.2: Unstandardised bivariate regression coefficients of models predicting well-being. Columns indicate predictors, and the two rows are the different well-being outcome variables.
4.3 RICLPM
Wrangle data to a format where lavaan model is easier to map to variable pairs: Wide format with different rows for outcomes (well-being) and predictors (hours, needs, motivations).
d_riclpm_long <- d %>%
select(
Game, pid, wid,
Hours, hours_est,
Intrinsic, Extrinsic,
Affect, `Life satisfaction`, AN, AP
) %>%
# Long format on well-being scores (outcomes)
pivot_longer(
c(Affect, `Life satisfaction`, AN, AP),
names_to = "y_var", values_to = "y"
) %>%
# Long format on other variables (predictors)
pivot_longer(
c(Hours, hours_est, Intrinsic, Extrinsic),
names_to = "x_var", values_to = "x"
)
d_riclpm_wide <- d_riclpm_long %>%
pivot_wider(names_from = wid, values_from = c(x, y), names_sep = "")
write_rds(d_riclpm_wide, here("Temp", "d_riclpm_wide.rds"))4.3.1 Cross-lagged scatterplots
Order <- tibble(
y_var = rep(c("Affect", "Life satisfaction"), each = 2),
wid = c(2, 3, 2, 3),
Panel = c(
"Affect[plain('[Wave 2]')]",
"Affect[plain('[Wave 3]')]",
"LS[plain('[Wave 2]')]",
"LS[plain('[Wave 3]')]"
),
Panel_label = fct_inorder(Panel)
)
d_riclpm_plots <- d_riclpm_long %>%
# Don't show AN and AP
filter(!(y_var %in% c("AN", "AP"))) %>%
# Take out hours per day over 3 for these plots
mutate(x = if_else(str_detect(x_var, "ours") & x > 3, NaN, x)) %>%
arrange(Game, pid, x_var, wid) %>%
group_by(Game, pid, x_var, y_var) %>%
mutate(lag_x = lag(x)) %>%
left_join(Order) %>%
ungroup() %>%
filter(wid > 1)
d_riclpm_plots %>%
filter(x_var == "Hours") %>%
ggplot(aes(lag_x, y)) +
scale_x_continuous(
"Hours played per day at previous wave",
breaks = pretty_breaks(3)
) +
scale_y_continuous(
"Well-being at current wave",
breaks = pretty_breaks()
) +
geom_point(size = .2, alpha = .2, shape = 1, col = col1) +
geom_smooth(
method = "gam", size = .4,
color = "black",
alpha = .33, show.legend = FALSE
) +
facet_grid(
Panel_label ~ Game,
scales = "free_y",
labeller = labeller(.rows = label_parsed)
) +
theme(
aspect.ratio = 1,
panel.grid = element_blank()
)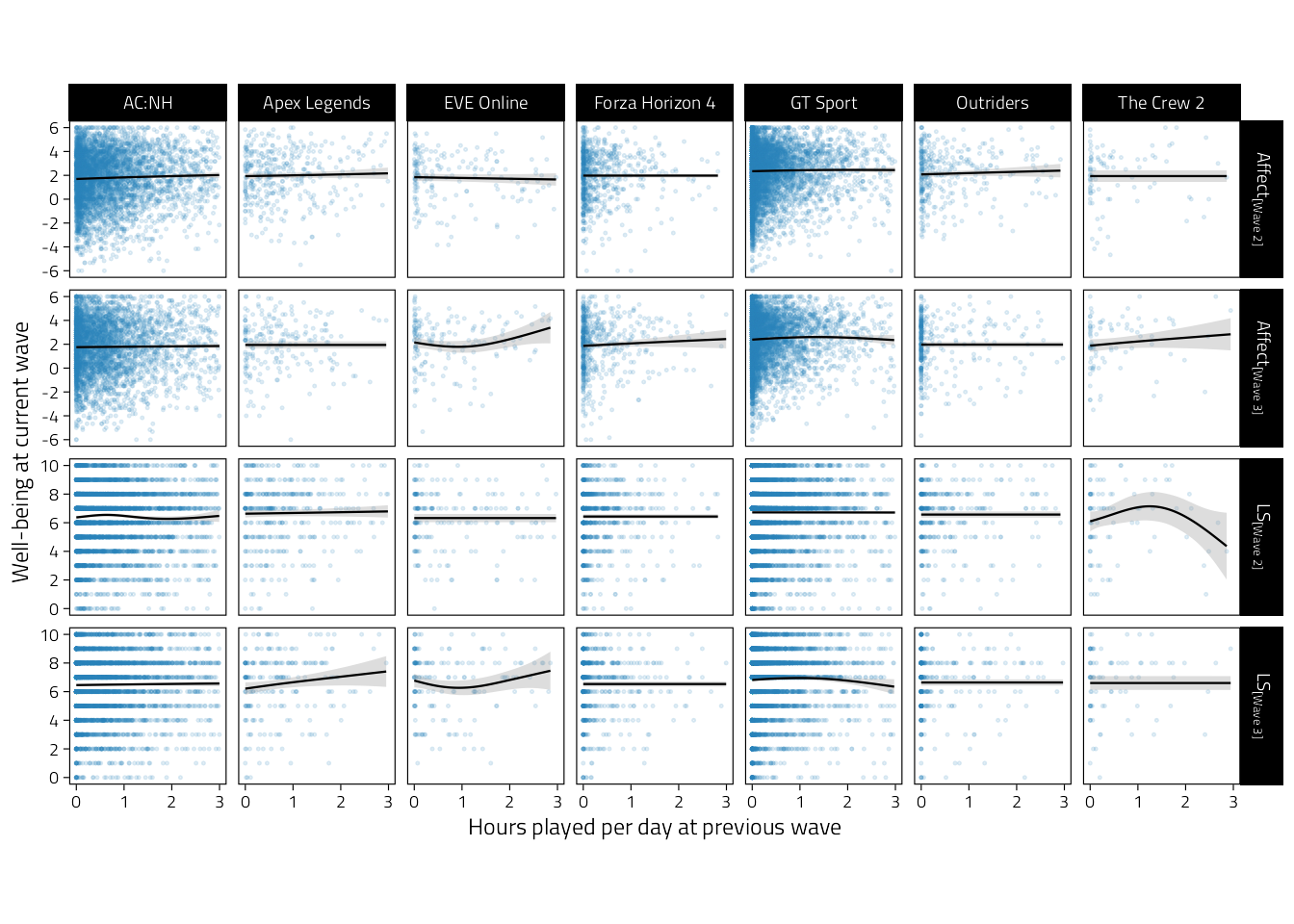
Figure 4.3: Scatterplots of well-being (rows indicate variables and waves) on average hours played per day during the previous wave. Regression lines are GAM fitted lines and 95%CIs.
# Plots with other predictors
p_tmp_0 <- last_plot() %+%
filter(d_riclpm_plots, x_var == "hours_est") +
scale_x_continuous(
"Estimated hours played per day at previous wave",
breaks = 0:3
)
p_tmp_0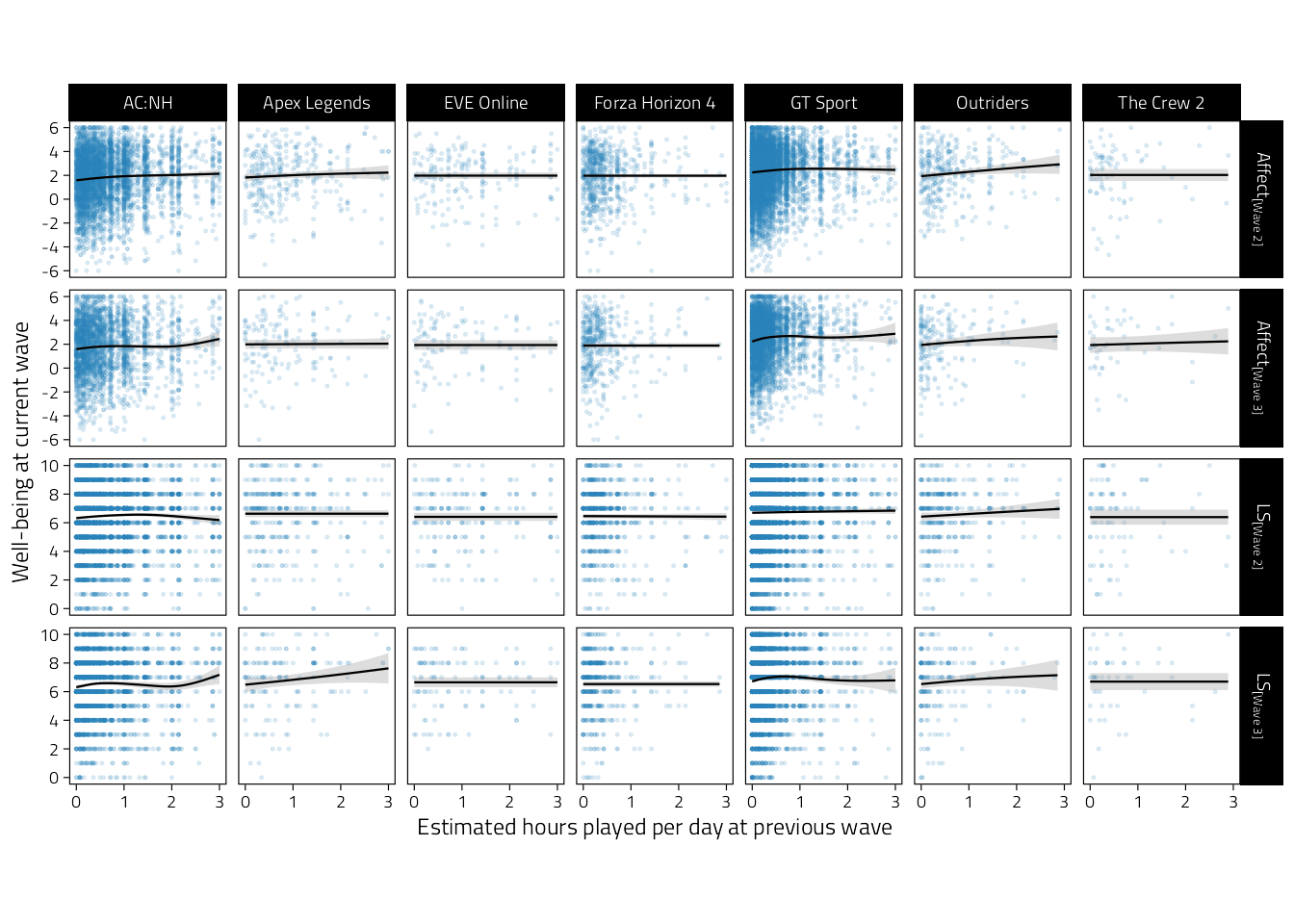
Figure 4.4: Scatterplots of well-being (rows indicate variables and waves) on estimated average hours played per day during the previous wave. Regression lines are GAM fitted lines and 95%CIs.
p_tmp_1 <- last_plot() %+%
filter(d_riclpm_plots, x_var == "Intrinsic") +
scale_x_continuous(
"Intrinsic motivation at previous wave",
breaks = pretty_breaks()
)
p_tmp_1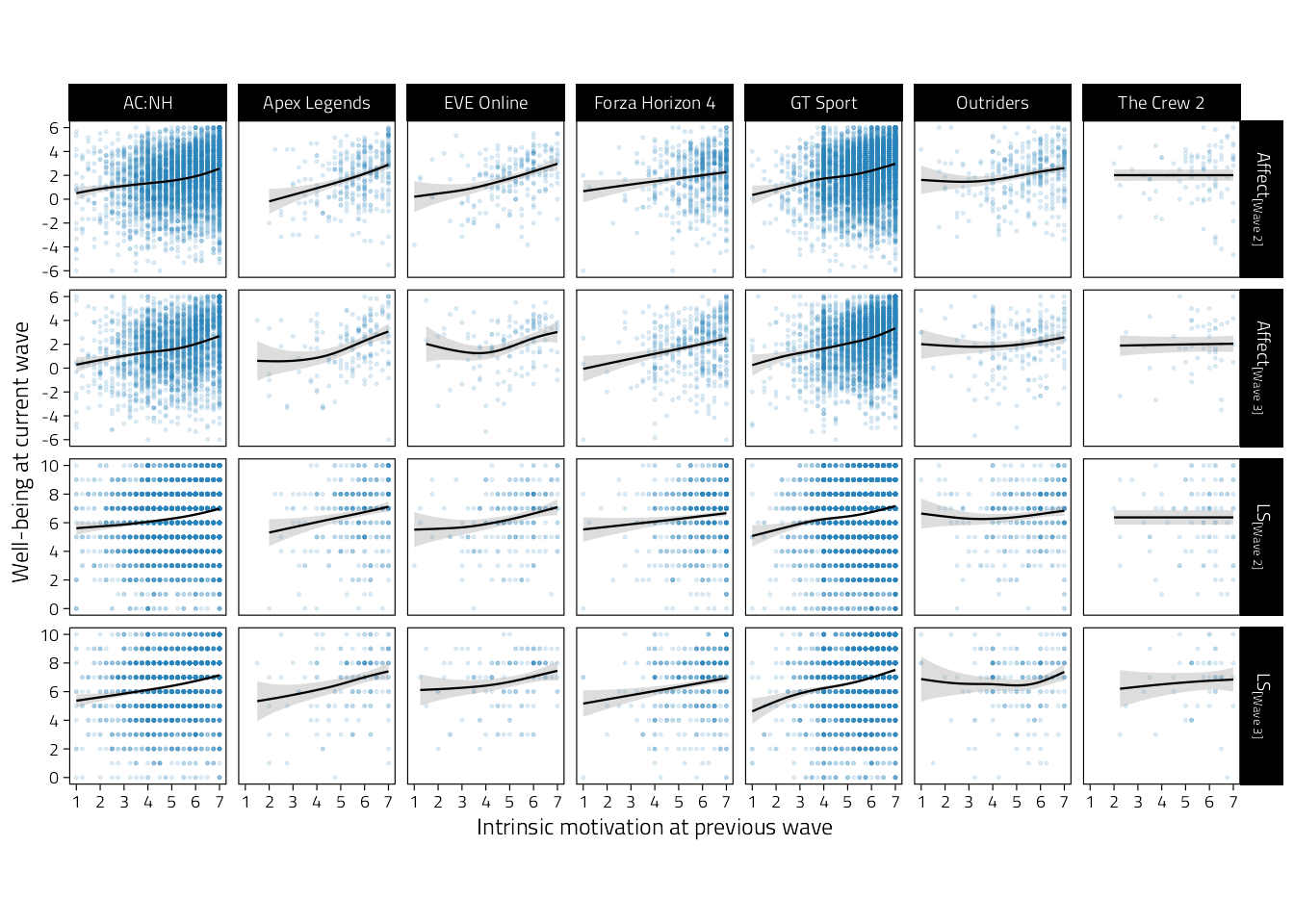
Figure 4.5: Scatterplots of well-being (rows indicate variables and waves) on intrinsic motivation during the previous wave. Regression lines are GAM fitted lines and 95%CIs.
p_tmp_2 <- last_plot() %+%
filter(d_riclpm_plots, x_var == "Extrinsic") +
scale_x_continuous(
"Extrinsic motivation at previous wave",
breaks = pretty_breaks()
)
p_tmp_2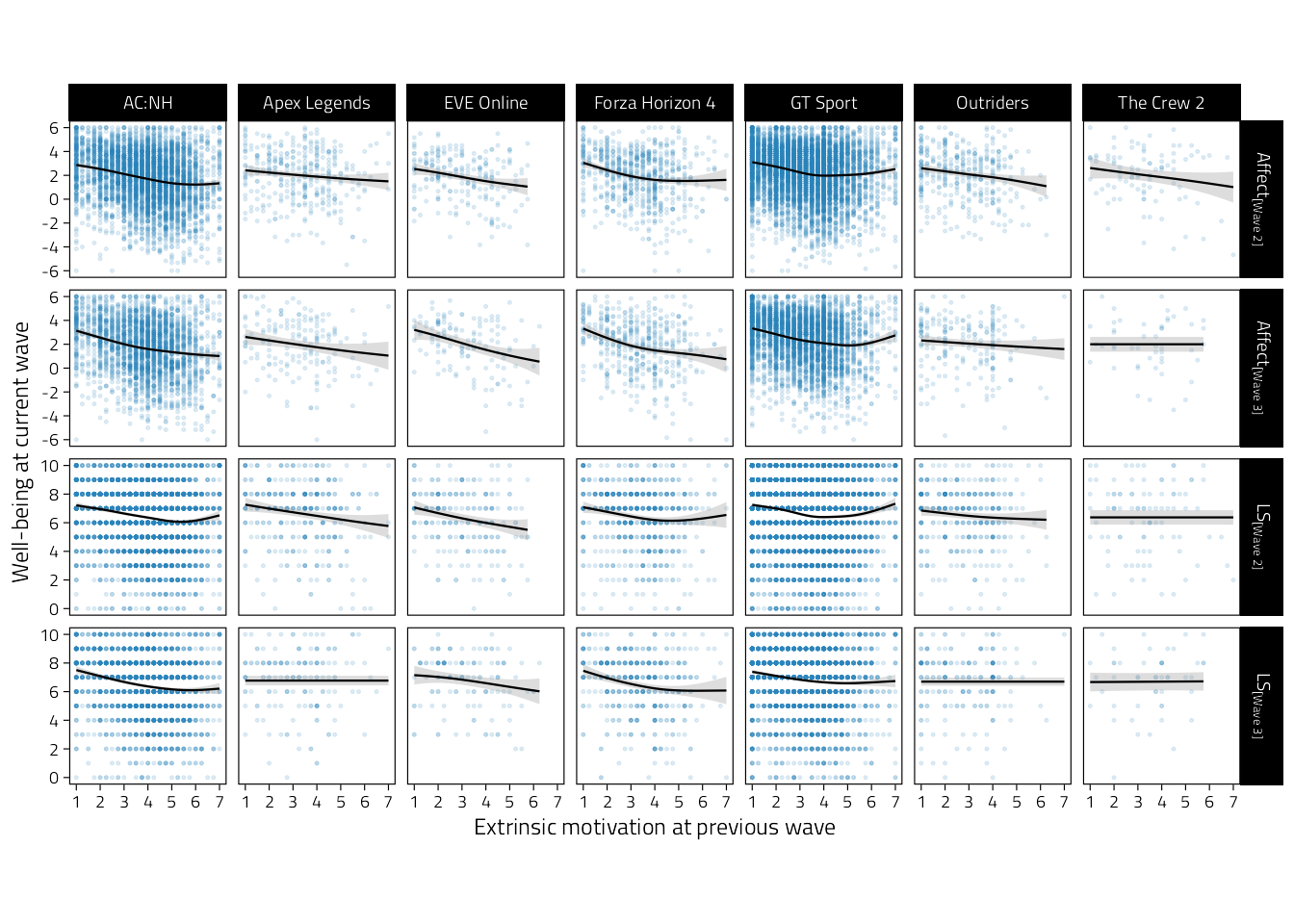
Figure 4.6: Scatterplots of well-being (rows indicate variables and waves) on extrinsic motivation during the previous wave. Regression lines are GAM fitted lines and 95%CIs.
p_tmp_1 / p_tmp_2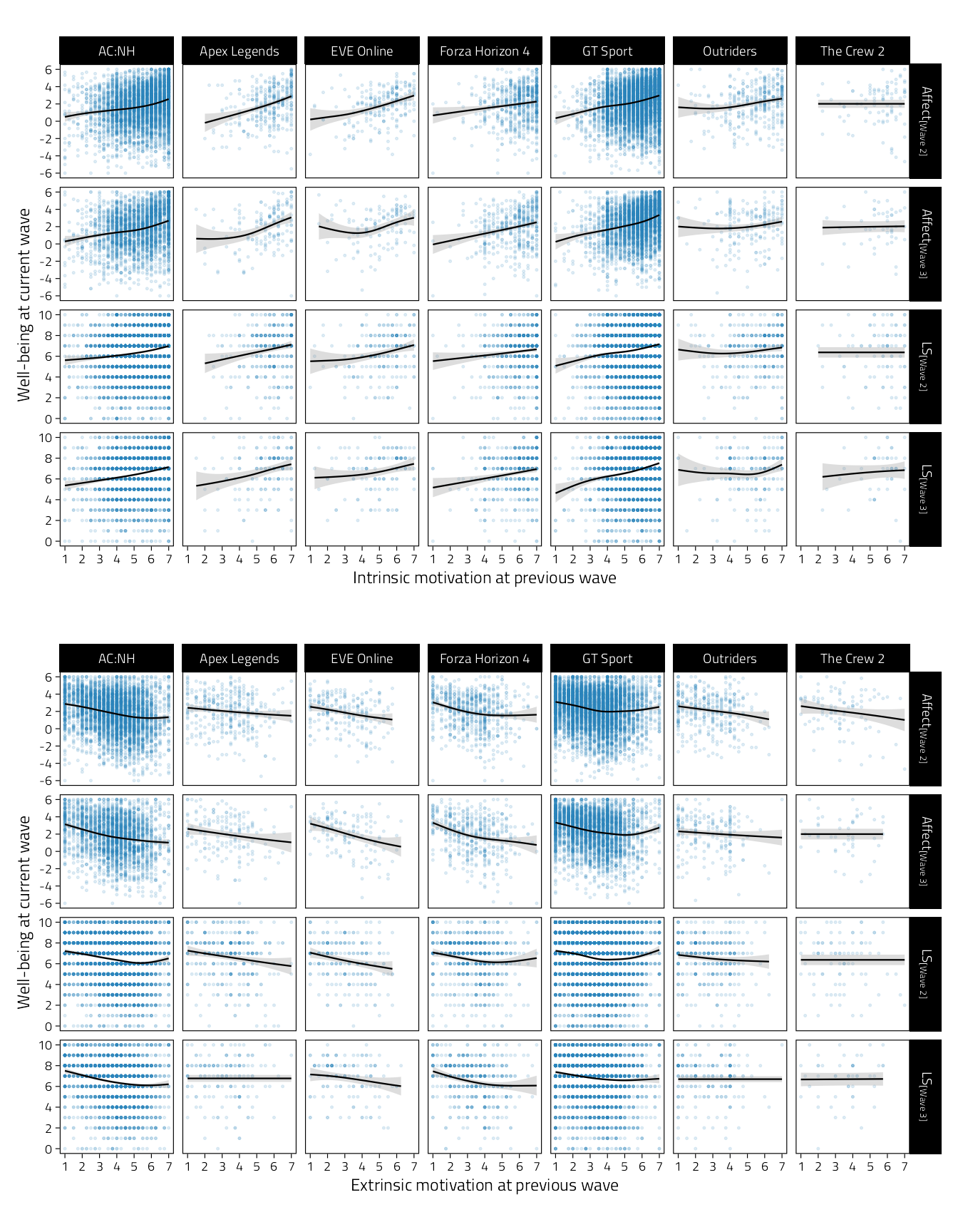
Figure 4.7: Scatterplots of well-being (rows indicate variables and waves) on intrinsic and extrinsic motivation during the previous wave. Regression lines are GAM fitted lines and 95%CIs.
4.3.2 Fit models
First fit RICLPM to all data. Separately to the two well-being outcomes, and each predictor (hours, subjective scales). Constrain (cross)lagged parameters
Syntax based on https://jeroendmulder.github.io/RI-CLPM/lavaan.html
riclpm_1 <- "
# Create between components (random intercepts)
RIx =~ 1*x1 + 1*x2 + 1*x3
RIy =~ 1*y1 + 1*y2 + 1*y3
# Create within-person centered variables
wx1 =~ 1*x1
wx2 =~ 1*x2
wx3 =~ 1*x3
wy1 =~ 1*y1
wy2 =~ 1*y2
wy3 =~ 1*y3
# Estimate the lagged effects between the within-person centered variables (constrained).
wx2 ~ bx*wx1 + gx*wy1
wy2 ~ gy*wx1 + by*wy1
wx3 ~ bx*wx2 + gx*wy2
wy3 ~ gy*wx2 + by*wy2
# Estimate the covariance between the within-person centered
# variables at the first wave.
wx1 ~~ wy1 # Covariance
# Estimate the covariances between the residuals of the
# within-person centered variables (the innovations).
wx2 ~~ wy2
wx3 ~~ wy3
# Estimate the variance and covariance of the random intercepts.
RIx ~~ RIx
RIy ~~ RIy
RIx ~~ RIy
# Estimate the (residual) variance of the within-person centered variables.
wx1 ~~ wx1 # Variances
wy1 ~~ wy1
wx2 ~~ wx2 # Residual variances
wy2 ~~ wy2
wx3 ~~ wx3
wy3 ~~ wy3
"
# This is the same but was suggested to not include autoregressions
riclpm_2 <- [1109 chars quoted with '"']
cluster_library(cluster, "purrr")
cluster_copy(cluster, c("riclpm_1", "riclpm_2"))
path <- "models/riclpm-pooled.rds"
if (!file.exists(path)) {
fit_riclpm_all <- d_riclpm_wide %>%
group_by(y_var, x_var) %>%
nest() %>%
crossing(
nesting(
lvn = c(riclpm_1, riclpm_2),
model = c("1", "2")
)
) %>%
partition(cluster) %>%
mutate(
fit = map2(
data, lvn,
~lavaan(
.y,
data = .x,
missing = "ml",
meanstructure = TRUE,
int.ov.free = TRUE
)
)
) %>%
collect() %>%
ungroup() %>%
select(-data, -lvn)
saveRDS(fit_riclpm_all, path)
} else {fit_riclpm_all <- readRDS(path)}Fit the above model separately to each game.
path <- "models/riclpm-games.rds"
if (!file.exists(path)) {
fit_riclpm_games <- d_riclpm_wide %>%
group_by(y_var, x_var, Game) %>%
nest() %>%
crossing(
nesting(
lvn = c(riclpm_1, riclpm_2),
model = c("1", "2")
)
) %>%
partition(cluster) %>%
mutate(
fit = map2(
data, lvn,
~lavaan(
.y,
data = .x,
missing = "ml",
meanstructure = TRUE,
int.ov.free = TRUE
)
)
) %>%
collect() %>%
ungroup() %>%
select(-data, -lvn)
saveRDS(fit_riclpm_games, path)
} else {fit_riclpm_games <- readRDS(path)}
get_lavaan_pars <- function(x) {
bind_rows(
parameterestimates(x) %>%
mutate(Type = "Unstandardized"),
standardizedsolution(x) %>%
rename(est = est.std) %>%
mutate(Type = "Standardized")
) %>%
as_tibble() %>%
unite("Parameter", c(lhs, op, rhs), sep = " ", remove = FALSE)
}
pars_riclpm <- bind_rows(
"Pooled" = fit_riclpm_all,
"Independent" = fit_riclpm_games,
.id = "Fit"
) %>%
mutate(pars = map(fit, get_lavaan_pars)) %>%
select(-fit)
# Take only parameters of interest
pars_riclpm <- pars_riclpm %>%
unnest(pars) %>%
filter(
(str_detect(Parameter, " ~ ") & !str_detect(Parameter, "3")) |
Parameter %in% c("RIx ~~ RIy", "wx1 ~~ wy1", "wx2 ~~ wy2", "wx3 ~~ wy3")
) %>%
select(-c(lhs:label, z))
pars_riclpm %>%
filter(Fit == "Pooled", model == "1") %>%
filter(
str_detect(Parameter, " ~ "),
Type == "Unstandardized"
) %>%
mutate(across(where(is.numeric), ~ format(round(.x, 2), nsmall = 2))) %>%
mutate(Result = str_glue("{est}, [{ci.lower}, {ci.upper}]")) %>%
select(y_var, x_var, Parameter, Result) %>%
pivot_wider(names_from = Parameter, values_from = Result) %>%
arrange(y_var, x_var) %>%
kbl(
caption = "RICLPM regression parameters from pooled model."
) %>%
kable_classic(full_width = FALSE, html_font = Font)| y_var | x_var | wx2 ~ wx1 | wx2 ~ wy1 | wy2 ~ wx1 | wy2 ~ wy1 |
|---|---|---|---|---|---|
| Affect | Extrinsic | 0.22, [ 0.18, 0.27] | -0.06, [-0.08, -0.04] | -0.12, [-0.17, -0.07] | 0.35, [ 0.32, 0.39] |
| Affect | Hours | 0.47, [ 0.46, 0.49] | 0.01, [ 0.00, 0.01] | -0.02, [-0.07, 0.03] | 0.37, [ 0.34, 0.41] |
| Affect | hours_est | 0.17, [ 0.14, 0.20] | 0.01, [ 0.00, 0.02] | -0.03, [-0.09, 0.03] | 0.37, [ 0.34, 0.41] |
| Affect | Intrinsic | 0.35, [ 0.32, 0.39] | 0.07, [ 0.06, 0.09] | 0.14, [ 0.09, 0.19] | 0.36, [ 0.32, 0.39] |
| AN | Extrinsic | 0.23, [ 0.18, 0.28] | 0.08, [ 0.04, 0.12] | 0.08, [ 0.05, 0.11] | 0.26, [ 0.22, 0.30] |
| AN | Hours | 0.47, [ 0.46, 0.49] | -0.01, [-0.02, 0.00] | -0.01, [-0.04, 0.03] | 0.28, [ 0.24, 0.32] |
| AN | hours_est | 0.17, [ 0.14, 0.21] | -0.02, [-0.04, 0.01] | 0.02, [-0.02, 0.05] | 0.28, [ 0.24, 0.32] |
| AN | Intrinsic | 0.37, [ 0.33, 0.40] | -0.11, [-0.14, -0.08] | -0.08, [-0.11, -0.05] | 0.28, [ 0.24, 0.32] |
| AP | Extrinsic | 0.23, [ 0.18, 0.28] | -0.08, [-0.11, -0.04] | -0.05, [-0.08, -0.02] | 0.30, [ 0.27, 0.34] |
| AP | Hours | 0.47, [ 0.46, 0.49] | 0.01, [ 0.00, 0.02] | -0.02, [-0.05, 0.02] | 0.31, [ 0.27, 0.35] |
| AP | hours_est | 0.17, [ 0.14, 0.20] | 0.01, [-0.01, 0.03] | -0.01, [-0.05, 0.03] | 0.31, [ 0.27, 0.35] |
| AP | Intrinsic | 0.36, [ 0.33, 0.40] | 0.10, [ 0.07, 0.13] | 0.09, [ 0.06, 0.12] | 0.30, [ 0.26, 0.33] |
| Life satisfaction | Extrinsic | 0.23, [ 0.18, 0.28] | -0.02, [-0.04, 0.00] | -0.07, [-0.14, 0.00] | 0.18, [ 0.14, 0.22] |
| Life satisfaction | Hours | 0.48, [ 0.46, 0.49] | 0.00, [-0.01, 0.00] | -0.06, [-0.12, 0.01] | 0.18, [ 0.14, 0.22] |
| Life satisfaction | hours_est | 0.17, [ 0.14, 0.21] | 0.01, [ 0.00, 0.02] | 0.03, [-0.05, 0.11] | 0.18, [ 0.14, 0.22] |
| Life satisfaction | Intrinsic | 0.38, [ 0.34, 0.41] | 0.04, [ 0.02, 0.05] | 0.18, [ 0.12, 0.24] | 0.18, [ 0.15, 0.22] |
4.3.3 Table of parameters
This table displays the lavaan estimates (game specific and pooled) for model 1 (full RICLPM) and 2 (RICLPM without autocorrelations).
# Interactive table so reader can find what they're looking for
pars_riclpm %>%
mutate(
pvalue = pvalue(pvalue),
across(c(model, x_var, y_var, Game, Parameter, Type, Fit), factor),
across(where(is.numeric), ~format(round(.x, 2), nsmall = 2)),
Estimate = str_glue("{est} [{ci.lower}, {ci.upper}]")
) %>%
select(
model, Fit, x_var, y_var, Game, Parameter, Type,
Estimate, pvalue
) %>%
datatable(
filter = "top",
class = "display",
rownames = FALSE
) %>%
formatStyle(TRUE, `font-size` = '12px')4.4 Meta-analyses
Number of observations/participants per model
fit_riclpm_games %>%
filter(model == 1) %>%
mutate(N = map_dbl(fit, nobs) %>% comma()) %>%
select(-fit, -model) %>%
pivot_wider(
names_from = x_var,
values_from = N,
names_glue = "{x_var} {.value}"
) %>%
rename(Outcome = y_var) %>%
arrange(Outcome, Game) %>%
kbl(caption = "Sample sizes per RICLPM") %>%
kable_classic(full_width = FALSE, html_font = Font)| Game | Outcome | Extrinsic N | Hours N | hours_est N | Intrinsic N |
|---|---|---|---|---|---|
| AC:NH | Affect | 13,485.0 | 13,646.0 | 13,477.0 | 13,487.0 |
| Apex Legends | Affect | 1,127.0 | 1,158.0 | 1,125.0 | 1,127.0 |
| EVE Online | Affect | 886.0 | 905.0 | 887.0 | 886.0 |
| Forza Horizon 4 | Affect | 1,951.0 | 1,981.0 | 1,953.0 | 1,951.0 |
| GT Sport | Affect | 18,964.0 | 19,258.0 | 18,975.0 | 18,972.0 |
| Outriders | Affect | 1,518.0 | 1,530.0 | 1,512.0 | 1,518.0 |
| The Crew 2 | Affect | 448.0 | 457.0 | 446.0 | 448.0 |
| AC:NH | AN | 13,487.0 | 13,646.0 | 13,479.0 | 13,489.0 |
| Apex Legends | AN | 1,127.0 | 1,158.0 | 1,125.0 | 1,127.0 |
| EVE Online | AN | 886.0 | 905.0 | 887.0 | 886.0 |
| Forza Horizon 4 | AN | 1,951.0 | 1,981.0 | 1,953.0 | 1,951.0 |
| GT Sport | AN | 18,966.0 | 19,258.0 | 18,976.0 | 18,973.0 |
| Outriders | AN | 1,518.0 | 1,530.0 | 1,512.0 | 1,518.0 |
| The Crew 2 | AN | 449.0 | 457.0 | 447.0 | 449.0 |
| AC:NH | AP | 13,487.0 | 13,646.0 | 13,480.0 | 13,488.0 |
| Apex Legends | AP | 1,127.0 | 1,158.0 | 1,125.0 | 1,127.0 |
| EVE Online | AP | 886.0 | 905.0 | 887.0 | 886.0 |
| Forza Horizon 4 | AP | 1,952.0 | 1,981.0 | 1,953.0 | 1,952.0 |
| GT Sport | AP | 18,976.0 | 19,258.0 | 18,980.0 | 18,983.0 |
| Outriders | AP | 1,518.0 | 1,530.0 | 1,512.0 | 1,518.0 |
| The Crew 2 | AP | 448.0 | 457.0 | 446.0 | 448.0 |
| AC:NH | Life satisfaction | 13,621.0 | 13,646.0 | 13,587.0 | 13,621.0 |
| Apex Legends | Life satisfaction | 1,152.0 | 1,158.0 | 1,148.0 | 1,152.0 |
| EVE Online | Life satisfaction | 904.0 | 905.0 | 897.0 | 904.0 |
| Forza Horizon 4 | Life satisfaction | 1,979.0 | 1,981.0 | 1,976.0 | 1,979.0 |
| GT Sport | Life satisfaction | 19,214.0 | 19,258.0 | 19,169.0 | 19,218.0 |
| Outriders | Life satisfaction | 1,526.0 | 1,530.0 | 1,519.0 | 1,526.0 |
| The Crew 2 | Life satisfaction | 452.0 | 457.0 | 454.0 | 452.0 |
Then conduct all meta-analyses (for each model, y_var-x_var pair, parameter of interest, and type of parameter (standardized, unstandardized)) of the game-specific RICLPM parameters.
# Select variables from RICLPM parameters to data for MA
d_ma <- pars_riclpm %>%
filter(
Fit == "Independent",
str_detect(Parameter, " ~ ") |
Parameter %in% c("wx1 ~~ wy1", "RIx ~~ RIy")
) %>%
select(
model, Type, y_var,
x_var, Game, Parameter,
est, se, ci.lower, ci.upper
)
path <- "models/all-meta-analyses.rds"
if (!file.exists(path)) {
cluster_copy(cluster, "get_ma_post")
cluster_library(
cluster,
c("brms", "purrr", "cmdstanr", "rstan", "bayestestR", "forcats")
)
fit_ma <- d_ma %>%
mutate(
name = str_glue("models/brm-{model}-{Type}-{y_var}-{x_var}-{Parameter}")
) %>%
group_by(model, x_var, y_var, Parameter, Type, name) %>%
nest() %>%
partition(cluster) %>%
mutate(
# Fit model
fit = map2(
data, name,
~brm(
bf(est | se(se) ~ 1 + (1 | Game)),
data = .x,
backend = "cmdstanr",
prior = prior(student_t(7, 0, 0.25), class = "sd", group = "Game"),
control = list(adapt_delta = .9999, max_treedepth = 15),
iter = 15000, warmup = 10000,
file = .y
)
),
# Table of posterior samples
posterior = map(fit, get_ma_post),
# Table of posterior summary
summary = map(
posterior,
~ describe_posterior(
.x,
centrality = "mean",
ci = 0.95,
ci_method = "eti",
test = "pd"
) %>%
rename(Game = Parameter) %>%
mutate(Game = fct_relevel(Game, "Average")) %>%
as_tibble()
),
#
sd = map(
fit,
~posterior_summary(.x, variable = "sd_Game__Intercept") %>%
as_tibble()
)
) %>%
collect() %>%
ungroup() %>%
select(-name, -fit)
saveRDS(fit_ma, path, compress = FALSE)
} else {fit_ma <- readRDS(path)}
# Arrange for tables etc
fit_ma <- fit_ma %>%
mutate(
model = factor(model, levels = 1:2, labels = c("AR", "No AR")),
Type = factor(Type, levels = c("Unstandardized", "Standardized")),
y_var = factor(
y_var, levels = c("Affect", "Life satisfaction", "AN", "AP")
),
x_var = factor(
x_var, levels = c("Hours", "Intrinsic", "Extrinsic", "hours_est")
),
Parameter = fct_relevel(Parameter, "wy2 ~ wx1")
) %>%
arrange(model, Type, y_var, x_var, Parameter)4.4.1 Table of parameters
All the resulting parameters are listed in the table below.
fit_ma %>%
select(model:Parameter, summary) %>%
unnest(summary) %>%
mutate(
across(where(is.numeric), ~format(round(.x, 2), nsmall = 2)),
Estimate = str_glue("{Mean} [{CI_low}, {CI_high}]")
) %>%
select(model, Type, y_var, x_var, Parameter, Game, Estimate, pd) %>%
datatable(
filter = "top",
class = "display",
rownames = FALSE
) %>%
formatStyle(TRUE, `font-size` = '12px')4.4.2 Hours <-> WB
Draw a forest plot of meta-analyses with hours. First, define informative labels and correct order for them, and load a function for drawing forest plots.
# Table of nice labels in proper order
Order <- distinct(fit_ma, x_var, y_var, Parameter) %>%
ungroup() %>%
# Meta-analyses only for regression parameters
filter(str_detect(Parameter, " ~ ")) %>%
mutate(
x_lab = case_when(
x_var == "Hours" ~ "Play",
x_var == "Intrinsic" ~ "Intrinsic",
x_var == "Extrinsic" ~ "Extrinsic"
),
y_lab = case_when(
y_var == "Affect" ~ "Affect",
y_var == "Life satisfaction" ~ "Life~satisfaction"
),
Panel_label = case_when(
Parameter == "wx2 ~ wx1" ~
str_glue('{x_lab}[plain("[t-1]")]%->%{x_lab}[plain("[t]")]'),
Parameter == "wy2 ~ wy1" ~
str_glue('{y_lab}[plain("[t-1]")]%->%{y_lab}[plain("[t]")]'),
Parameter == "wy2 ~ wx1" ~
str_glue('{x_lab}[plain("[t-1]")]%->%{y_lab}[plain("[t]")]'),
Parameter == "wx2 ~ wy1" ~
str_glue('{y_lab}[plain("[t-1]")]%->%{x_lab}[plain("[t]")]')
)
) %>%
mutate(
Parameter_order = factor(
Parameter,
levels = c("wy2 ~ wx1", "wx2 ~ wy1")
)
) %>%
arrange(Parameter_order, y_var) %>%
mutate(Panel_label = fct_inorder(Panel_label))
# Only these are needed
forest_plot_data <- fit_ma %>%
filter(
model == "AR",
Type == "Unstandardized",
y_var %in% c("Affect", "Life satisfaction"),
x_var %in% c("Hours", "Intrinsic", "Extrinsic"),
Parameter %in% c("wy2 ~ wx1", "wx2 ~ wy1")
) %>%
left_join(Order) %>%
select(-model, -Type)
forest_plot_data %>%
filter(x_var == "Hours") %>%
forest_plot()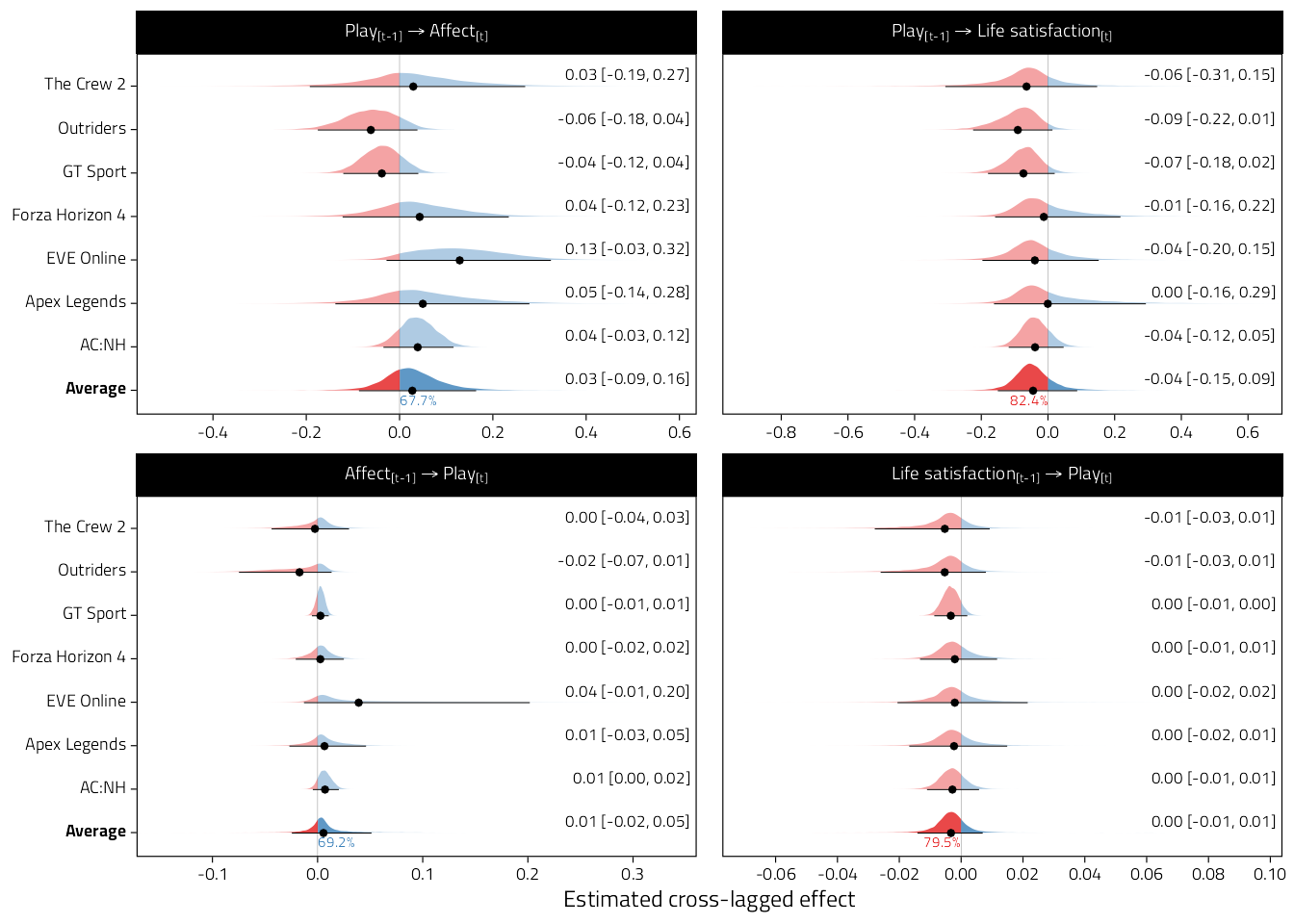
Figure 4.8: Plot
# Also see in comparison to lavaan model estimates
forest_plot_data %>%
filter(x_var == "Hours") %>%
forest_plot(lavaan = TRUE)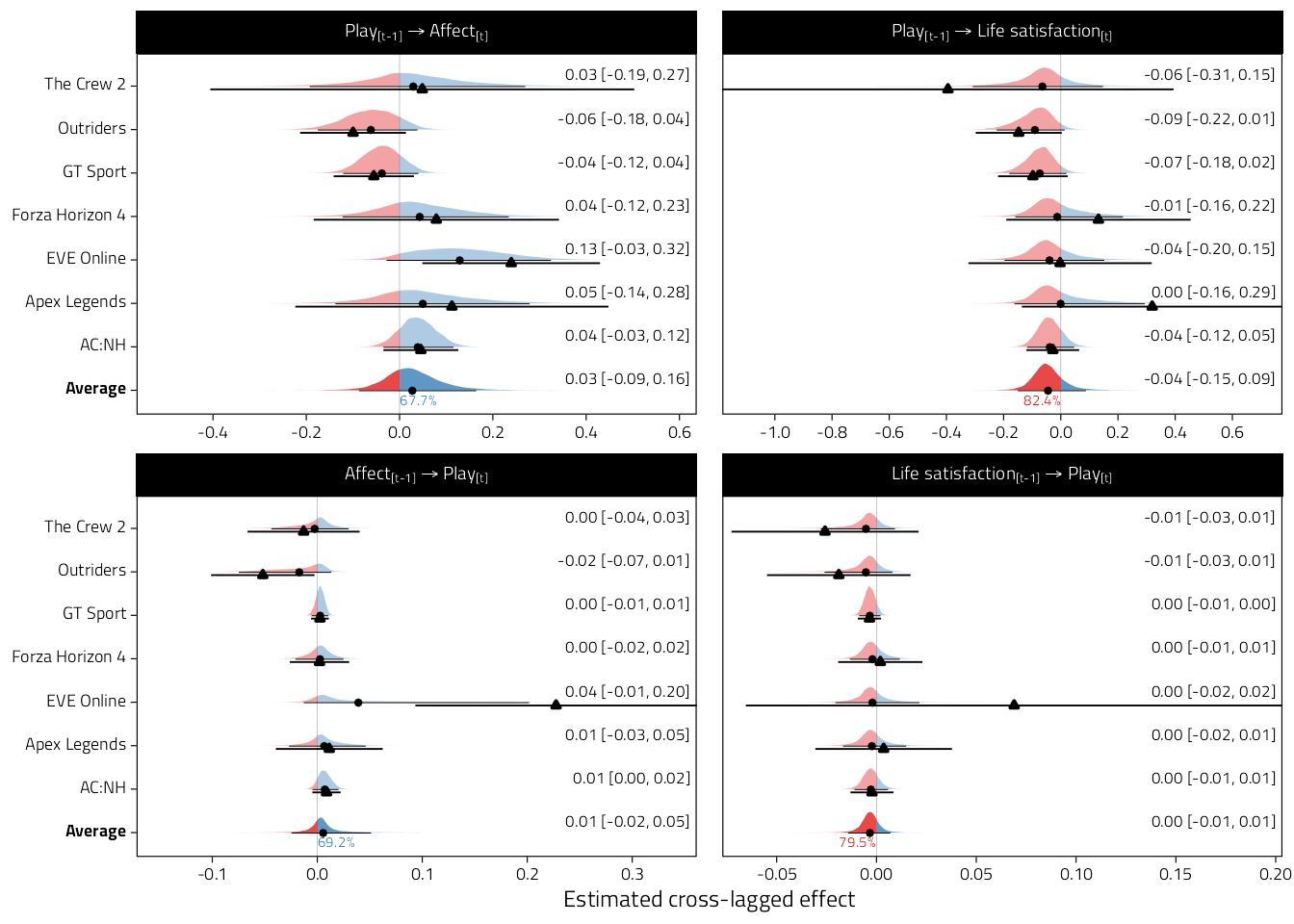
Figure 4.9: Figure as above but also displaying the underlying lavaan fits’ point estimates and 95%CIs.
4.4.3 Experiences <-> WB
a <- forest_plot_data %>%
filter(x_var %in% c("Intrinsic"), Parameter == "wy2 ~ wx1") %>%
forest_plot() +
theme(axis.title.x = element_blank())
b <- forest_plot_data %>%
filter(x_var %in% c("Extrinsic"), Parameter == "wy2 ~ wx1") %>%
forest_plot()
a/b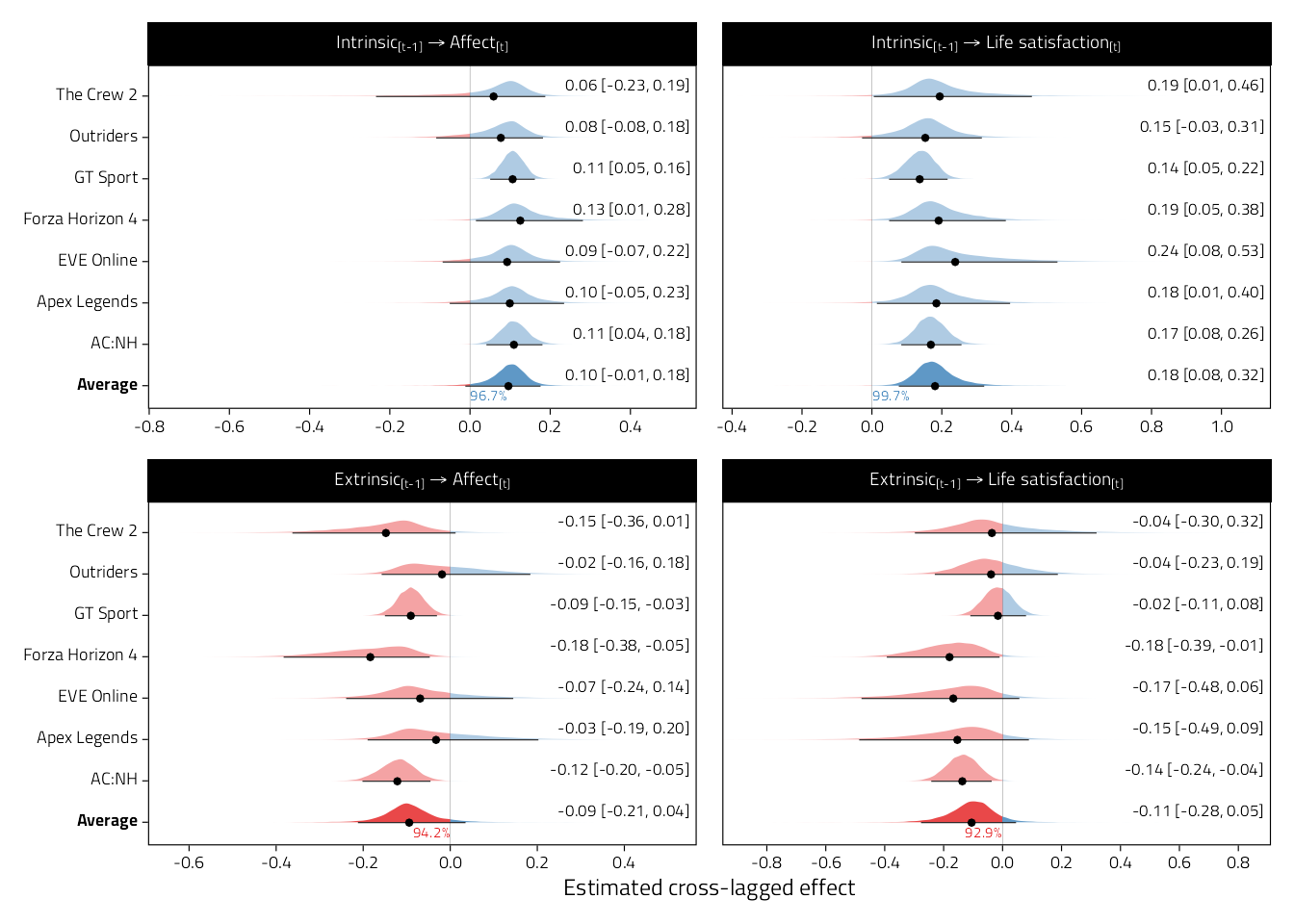
Figure 4.10: Forest plots of meta-analytic raw cross-lagged regression coefficients from models examining motivations and well-being (motivations to well-being). Shaded curves indicate approximate posterior densities, which are numerically summarised by means and 95%CIs in the right margin. Numbers below average effects indicate posterior probabilities of direction.
4.4.4 Some post-hoc calculations
The model parameters report effects of one-unit (e.g. one hour per day) changes in the predictor on the outcome. We also expand on this by first asking what is the effect of an average deviation in a predictor on the outcome. That is, for example, we calculate the average range of players’ average daily hours played, and then answer what the effect of an average player moving from their smallest to greatest amount of play would be on well-being.
# Average ranges of predictors
avg_ranges <- d %>%
# Calculate range for everyone (returns -Inf if all values missing)
group_by(Game, pid) %>%
summarise(
across(
c(Intrinsic, Extrinsic, Hours, hours_est),
.fns = ~max(.x, na.rm = TRUE) - min(.x, na.rm = TRUE)
)
) %>%
ungroup() %>%
# Calculate grand mean range excluding -Inf values
summarise(
across(
Intrinsic:hours_est,
~mean(if_else(.x==-Inf, NaN, .x), na.rm = TRUE)
)
) %>%
pivot_longer(
everything(),
names_to = "x_var",
values_to = "Mean_range"
)
fit_ma %>%
filter(
model == "AR",
Type == "Unstandardized",
Parameter == "wy2 ~ wx1",
x_var != "hours_est",
y_var %in% c("Affect", "Life satisfaction")
) %>%
left_join(avg_ranges) %>%
mutate(
hypothesis = map2(
posterior,
Mean_range,
~ hypothesis(.x, str_glue("Average * {.y} = 0")) %>% .[[1]]
)
) %>%
unnest(hypothesis) %>%
select(x_var, y_var, Mean_range, Parameter, Estimate:CI.Upper) %>%
kbl(digits = 3, caption = "ES of average hours per week on WB") %>%
kable_classic(full_width = FALSE, html_font = Font)| x_var | y_var | Mean_range | Parameter | Estimate | Est.Error | CI.Lower | CI.Upper |
|---|---|---|---|---|---|---|---|
| Hours | Affect | 0.479 | wy2 ~ wx1 | 0.013 | 0.030 | -0.042 | 0.079 |
| Intrinsic | Affect | 0.364 | wy2 ~ wx1 | 0.035 | 0.017 | -0.004 | 0.064 |
| Extrinsic | Affect | 0.408 | wy2 ~ wx1 | -0.038 | 0.024 | -0.086 | 0.014 |
| Hours | Life satisfaction | 0.479 | wy2 ~ wx1 | -0.021 | 0.028 | -0.072 | 0.042 |
| Intrinsic | Life satisfaction | 0.364 | wy2 ~ wx1 | 0.066 | 0.022 | 0.028 | 0.117 |
| Extrinsic | Life satisfaction | 0.408 | wy2 ~ wx1 | -0.043 | 0.033 | -0.113 | 0.019 |
We then turn to a second question, how much would the average player need to increase their play to result in a subjectively noticeable effect. Anvari and Lakens estimated that a 2% movement might be a lower limit to a subjectively noticeable change in well-being using a similar measurement scale.
We calculate, for each predictor and outcome, how much the predictor would need to change in order to elicit a change in the outcome greater than the subjective threshold. e.g. How many more hours would one need to play to “feel it”.
# Subjective threshold for each outcome
subjective_thresholds <- tibble(
y_var = c("Affect", "Life satisfaction"),
Threshold = 0.02 * c(13, 11)
)
# Table of effects and thresholds
fit_ma %>%
filter(
model == "AR",
Type == "Unstandardized",
Parameter == "wy2 ~ wx1",
x_var != "hours_est",
y_var %in% c("Affect", "Life satisfaction")
) %>%
select(x_var, y_var, summary) %>%
arrange(x_var, y_var) %>%
unnest(summary) %>%
select(x_var, y_var, Game, Effect = Mean) %>%
filter(Game == "Average") %>%
left_join(subjective_thresholds) %>%
mutate(`Change needed` = Threshold / abs(Effect)) %>%
kbl(
digits = 2,
caption = "Estimated required change in X to elicit a subjectively noticeable change in well-being outcomes"
) %>%
kable_classic(full_width = FALSE, html_font = Font)| x_var | y_var | Game | Effect | Threshold | Change needed |
|---|---|---|---|---|---|
| Hours | Affect | Average | 0.03 | 0.26 | 9.50 |
| Hours | Life satisfaction | Average | -0.04 | 0.22 | 4.93 |
| Intrinsic | Affect | Average | 0.10 | 0.26 | 2.72 |
| Intrinsic | Life satisfaction | Average | 0.18 | 0.22 | 1.22 |
| Extrinsic | Affect | Average | -0.09 | 0.26 | 2.76 |
| Extrinsic | Life satisfaction | Average | -0.11 | 0.22 | 2.09 |
How much of posterior density is within ROPE defined by A&L subjectively noticeable limits?
# Table of percentages of effects' posterior distributions with ROPE (as defined by thresholds for being subjectively noticed)
fit_ma %>%
filter(
model == "AR",
Type == "Unstandardized",
Parameter == "wy2 ~ wx1",
x_var != "hours_est",
y_var %in% c("Affect", "Life satisfaction")
) %>%
left_join(subjective_thresholds) %>%
mutate(
out = map2(
posterior,
Threshold,
~ describe_posterior(.x, rope_range = c(-.y, .y), rope_ci = 1) %>%
rename(Game = Parameter) %>%
filter(Game == "Average")
)
) %>%
select(x_var, y_var, out, Threshold) %>%
unnest(out) %>%
select(x_var, y_var, Threshold, Game, CI_low, CI_high, ROPE_Percentage) %>%
mutate(ROPE_Percentage = percent(ROPE_Percentage, .1)) %>%
kbl(
digits = 2,
caption = "ROPE percentages below threshold"
) %>%
kable_classic(full_width = FALSE, html_font = Font)| x_var | y_var | Threshold | Game | CI_low | CI_high | ROPE_Percentage |
|---|---|---|---|---|---|---|
| Hours | Affect | 0.26 | Average | -0.09 | 0.16 | 99.7% |
| Intrinsic | Affect | 0.26 | Average | 0.00 | 0.18 | 99.8% |
| Extrinsic | Affect | 0.26 | Average | -0.22 | 0.03 | 99.1% |
| Hours | Life satisfaction | 0.22 | Average | -0.16 | 0.08 | 99.5% |
| Intrinsic | Life satisfaction | 0.22 | Average | 0.07 | 0.31 | 79.2% |
| Extrinsic | Life satisfaction | 0.22 | Average | -0.27 | 0.05 | 92.8% |
4.5 Subjective time
Comparing subjective hours estimates to objective play behaviour is complicated by the fact that a missing value in objective behaviour indicates zero hours of play, whereas a missing value in estimated time can indicate either an estimated lack of play, or a missing response (e.g. player declined to respond to this item). Therefore any comparison here is done only on individuals who provided subjective estimate data.
Histograms of hours played (behaviour: top, strong; estimated: bottom, light). Only person-waves with both variables present are included.
tmp <- d %>%
drop_na(Hours, hours_est) %>%
group_by(Game, pid, Wave) %>%
summarise(
Hours = sum(Hours, na.rm = T),
hours_est = sum(hours_est, na.rm = T)
) %>%
group_by(Game, Wave) %>%
summarise(
across(c(Hours, hours_est), .fns = list(m = mean, se = ~ sd(.x) / sqrt(length(.x))))
)
d %>%
select(Game, pid, Wave, Hours, hours_est) %>%
drop_na(Hours, hours_est) %>%
filter(Hours < 3, hours_est < 3) %>%
ggplot(aes()) +
scale_y_continuous(
"Normalised count",
breaks = pretty_breaks()
) +
scale_x_continuous(breaks = pretty_breaks()) +
geom_histogram(
aes(x = Hours, y = stat(ncount)),
bins = 30, fill = col1, col = "white", alpha = .75
) +
geom_histogram(
aes(x = hours_est, y = stat(ncount) * -1),
bins = 30, fill = col1, col = "white", alpha = .5
) +
geom_pointrangeh(
data = tmp, shape = 25, fill = col1, alpha = 1,
aes(
x = Hours_m,
xmin = Hours_m - Hours_se,
xmax = Hours_m + Hours_se,
y = .075,
)
) +
geom_pointrangeh(
data = tmp, shape = 24, fill = col1, alpha = .75,
aes(
x = hours_est_m,
xmin = hours_est_m - hours_est_se,
xmax = hours_est_m + hours_est_se,
y = -.075
)
) +
facet_grid(Game ~ Wave, scales = "free")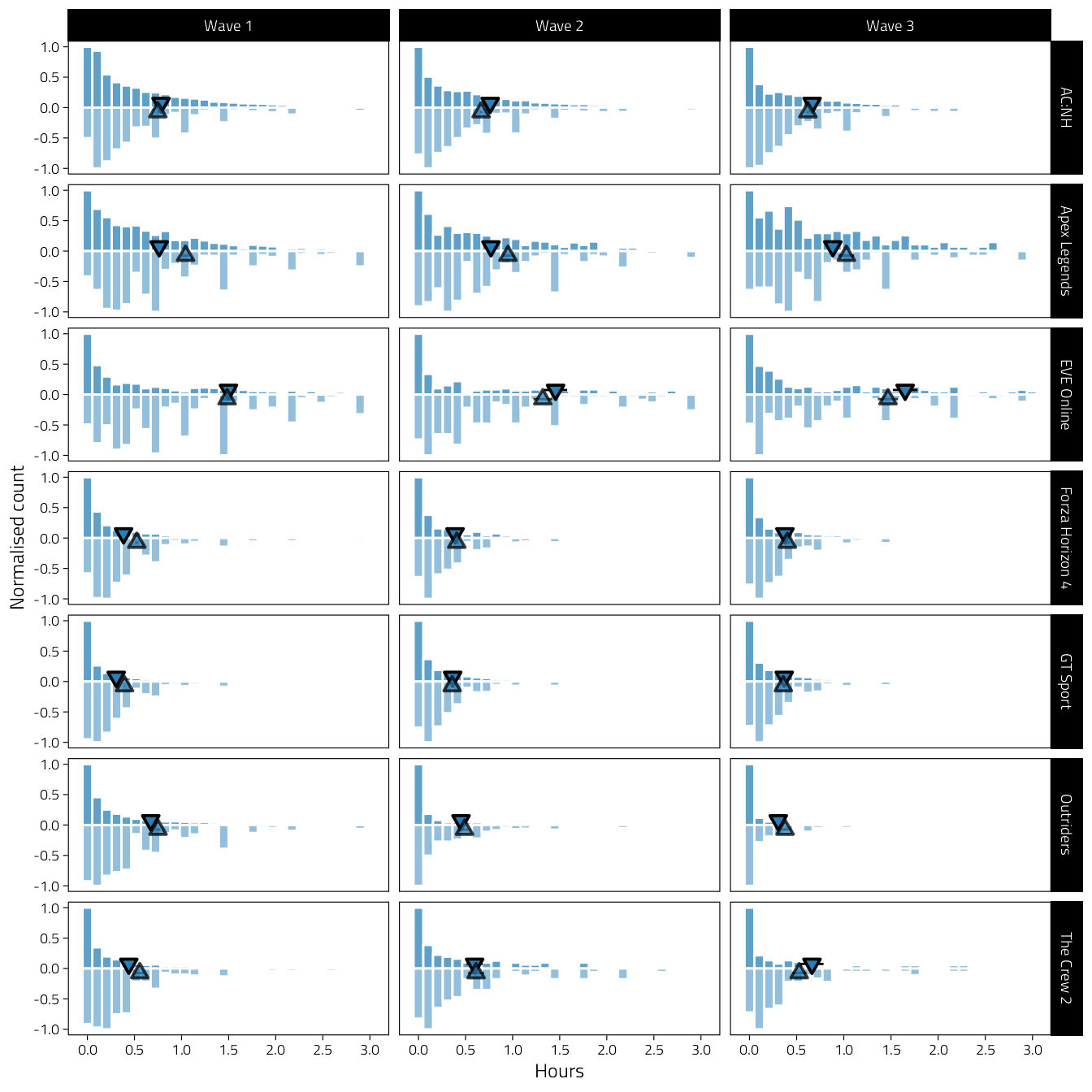
Figure 4.11: Bivariate histograms of average hours played per day by wave (columns) and game (rows). Histograms above zero, with strong colour, reflect behavioural telemetry log data. Histograms below zero, in light colour, indicate subjective estimates of play. Histogram heights are normalized counts. Small triangles indicate means.
A scatterplot of subjective on objective time. Only person-waves with both variables present are included.
d %>%
drop_na(Hours, hours_est) %>%
filter(Hours < 3, hours_est < 3) %>%
select(Game, pid, Wave, Objective = Hours, Subjective = hours_est) %>%
ggplot(aes(Objective, Subjective)) +
scale_x_continuous(
"Hours played per day (behaviour)",
breaks = c(0, 1, 2, 3), labels = c(0, 1, 2, 3)
) +
scale_y_continuous(
"Hours played per day (estimate)",
breaks = pretty_breaks()
) +
geom_point(size = .2, alpha = .2, shape = 1, col = col1) +
geom_abline(intercept = 0, slope = 1, lty = 2, size = .25) +
stat_centroid() +
geom_smooth(
method = "lm", size = .4,
color = "black",
alpha = .33, show.legend = FALSE
) +
facet_grid(Wave ~ Game) +
theme(aspect.ratio = 1)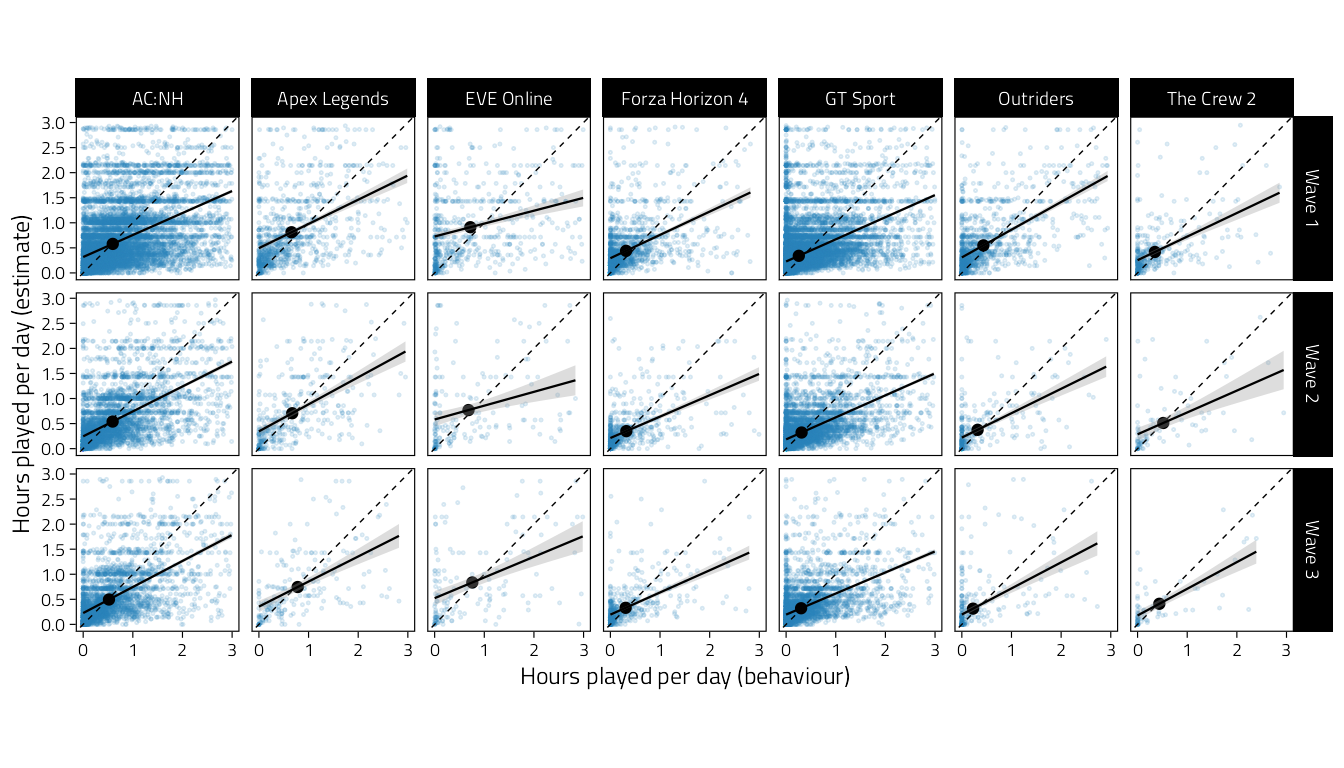
Figure 4.12: Scatterplots of subjectively estimated average hours of daily play on objective behavioural hours played. Dashed line indicates identity, solid lines are simple regression lines. Small blue points are individual participants, and solid dark points are bivariate sample means.
Wave-person differences. Difference indicates estimate - behaviour, so positive numbers indicate overestimates.
d %>%
select(Game, pid, Wave, Hours, hours_est) %>%
mutate(Difference = hours_est - Hours) %>%
filter(Hours < 3, hours_est < 3) %>%
drop_na(Difference) %>%
ggplot(aes(Difference, Game)) +
scale_x_continuous(
"Estimation error (hours/day)",
breaks = pretty_breaks()
) +
coord_cartesian(xlim = c(-0.5, 0.5)) +
geom_vline(xintercept = 0, lty = 2, size = .25) +
stat_halfeye(point_interval = NULL) +
stat_summaryh(fun.data = mean_cl_boot_h) +
facet_wrap("Wave", ncol = 1)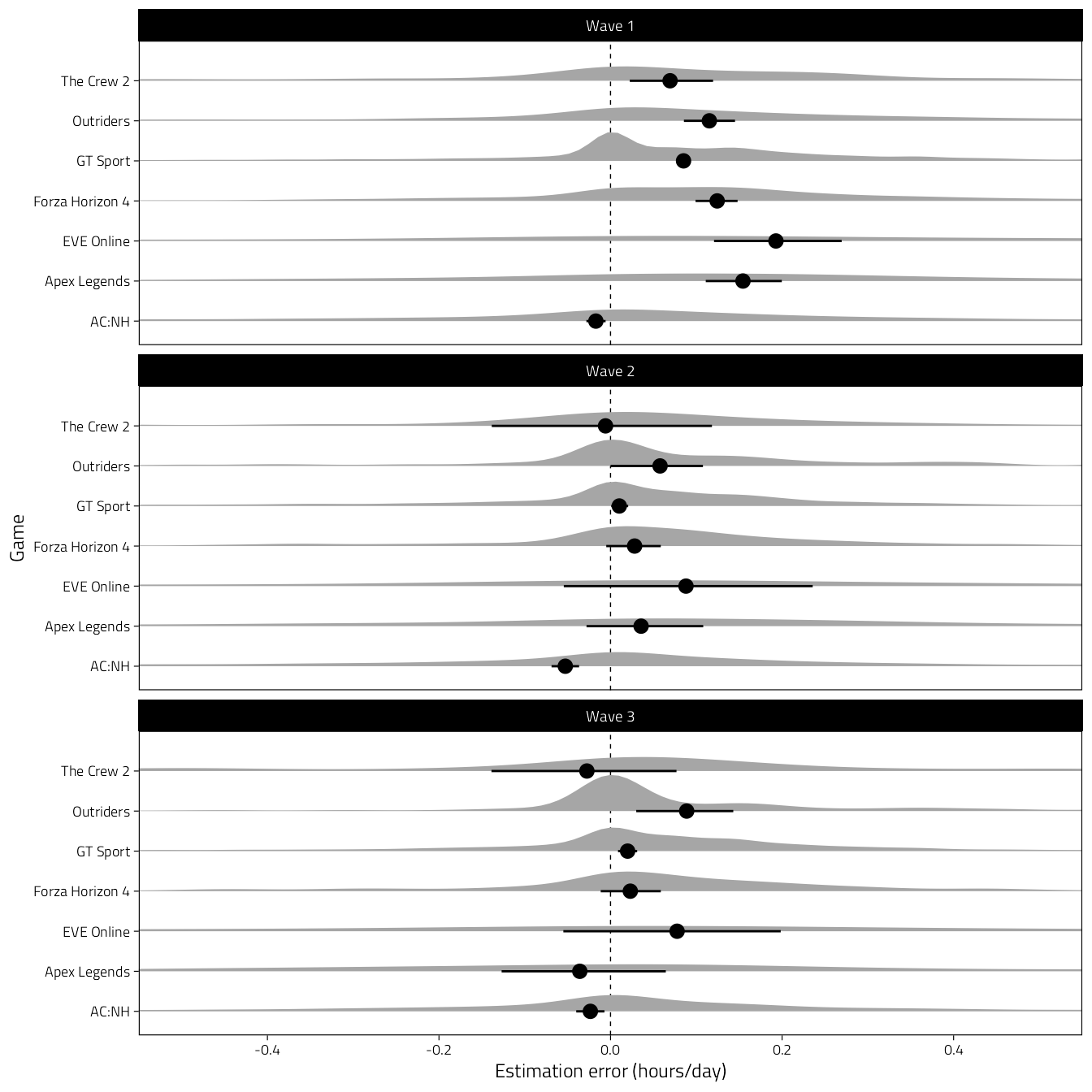
Figure 4.13: Density curves of differences between subjectively estimated and objectively logged average daily play time.
# Overall confidence interval for estimation error
d %>%
select(Game, pid, Wave, Hours, hours_est) %>%
mutate(Difference = hours_est - Hours) %>%
filter(Hours < 3, hours_est < 3) %>%
drop_na(Difference) %>%
group_by(Wave) %>%
summarise(mean_cl_boot(Difference)) %>%
kbl(digits = 2, caption = "Estimation errors (Estimated - Behaviour)") %>%
kable_classic(full_width = FALSE, html_font = Font)| Wave | y | ymin | ymax |
|---|---|---|---|
| Wave 1 | 0.06 | 0.05 | 0.06 |
| Wave 2 | -0.01 | -0.01 | 0.00 |
| Wave 3 | 0.01 | 0.00 | 0.02 |
4.5.1 Meta-analysis
fit_ma %>%
filter(
model == "AR",
Type == "Unstandardized",
x_var == "Hours",
y_var %in% c("Affect", "Life satisfaction"),
Parameter == "wy2 ~ wx1"
) %>%
select(-posterior) %>%
unnest(summary) %>%
mutate(
pd = percent(pd, .1),
across(where(is.numeric), ~format(round(.x, 2), nsmall = 2)),
Estimate = str_glue("{Mean} [{CI_low}, {CI_high}]")
) %>%
select(y_var, Game, Estimate, pd) %>%
kbl(caption = "Meta-analytic parameters of subjective time on WB") %>%
row_spec(c(8, 16), TRUE) %>%
kable_classic(full_width = FALSE, html_font = Font)| y_var | Game | Estimate | pd |
|---|---|---|---|
| Affect | AC:NH | 0.04 [-0.03, 0.12] | 84.5% |
| Affect | Apex Legends | 0.05 [-0.14, 0.28] | 68.5% |
| Affect | EVE Online | 0.13 [-0.03, 0.32] | 92.9% |
| Affect | Forza Horizon 4 | 0.04 [-0.12, 0.23] | 68.3% |
| Affect | GT Sport | -0.04 [-0.12, 0.04] | 81.6% |
| Affect | Outriders | -0.06 [-0.18, 0.04] | 86.5% |
| Affect | The Crew 2 | 0.03 [-0.19, 0.27] | 60.4% |
| Affect | Average | 0.03 [-0.09, 0.16] | 67.7% |
| Life satisfaction | AC:NH | -0.04 [-0.12, 0.05] | 82.3% |
| Life satisfaction | Apex Legends | 0.00 [-0.16, 0.29] | 62.2% |
| Life satisfaction | EVE Online | -0.04 [-0.20, 0.15] | 74.4% |
| Life satisfaction | Forza Horizon 4 | -0.01 [-0.16, 0.22] | 64.8% |
| Life satisfaction | GT Sport | -0.07 [-0.18, 0.02] | 93.9% |
| Life satisfaction | Outriders | -0.09 [-0.22, 0.01] | 95.4% |
| Life satisfaction | The Crew 2 | -0.06 [-0.31, 0.15] | 79.9% |
| Life satisfaction | Average | -0.04 [-0.15, 0.09] | 82.4% |
4.6 Variability
fit_ma %>%
filter(
model == "AR",
Type == "Unstandardized",
y_var %in% c("Affect", "Life satisfaction"),
x_var == "Hours",
Parameter == "wy2 ~ wx1"
) %>%
select(y_var, sd) %>%
unnest(sd) %>%
kbl(
caption = "Between game standard deviations of hours -> WB", digits = 2
) %>%
kable_classic(full_width = FALSE, html_font = Font)| y_var | Estimate | Est.Error | Q2.5 | Q97.5 |
|---|---|---|---|---|
| Affect | 0.11 | 0.06 | 0.01 | 0.27 |
| Life satisfaction | 0.08 | 0.07 | 0.00 | 0.26 |
4.7 System information
## R version 4.1.3 (2022-03-10)
## Platform: aarch64-apple-darwin20 (64-bit)
## Running under: macOS Monterey 12.3
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/4.1-arm64/Resources/lib/libRblas.0.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/4.1-arm64/Resources/lib/libRlapack.dylib
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## attached base packages:
## [1] stats graphics grDevices datasets utils methods base
##
## other attached packages:
## [1] broom.mixed_0.2.7 lme4_1.1-27.1 Matrix_1.3-4 cmdstanr_0.4.0
## [5] janitor_2.1.0 tidybayes_3.0.1 forcats_0.5.1 stringr_1.4.0
## [9] dplyr_1.0.7 purrr_0.3.4 readr_2.0.2 tidyr_1.1.4
## [13] tibble_3.1.5 tidyverse_1.3.1 multidplyr_0.1.0 lavaan_0.6-9
## [17] patchwork_1.1.1 cachem_1.0.6 memoise_2.0.1 ggdist_3.0.0
## [21] ggstance_0.3.5 ggtext_0.1.1 here_1.0.1 ragg_1.1.3
## [25] brms_2.16.1 Rcpp_1.0.7 bayestestR_0.11.0 ggpp_0.4.2
## [29] kableExtra_1.3.4 DT_0.19 scales_1.1.1 knitr_1.36
## [33] ggplot2_3.3.5
##
## loaded via a namespace (and not attached):
## [1] utf8_1.2.2 tidyselect_1.1.1 htmlwidgets_1.5.4
## [4] grid_4.1.3 munsell_0.5.0 codetools_0.2-18
## [7] miniUI_0.1.1.1 withr_2.4.2 Brobdingnag_1.2-6
## [10] colorspace_2.0-2 highr_0.9 rstudioapi_0.13
## [13] stats4_4.1.3 bayesplot_1.8.1 emmeans_1.7.2
## [16] rstan_2.21.2 mnormt_2.0.2 farver_2.1.0
## [19] datawizard_0.2.1 bridgesampling_1.1-2 rprojroot_2.0.2
## [22] coda_0.19-4 vctrs_0.3.8 generics_0.1.0
## [25] xfun_0.26 R6_2.5.1 markdown_1.1
## [28] gamm4_0.2-6 projpred_2.0.2 assertthat_0.2.1
## [31] promises_1.2.0.1 nnet_7.3-16 gtable_0.3.0
## [34] downlit_0.2.1 processx_3.5.2 rlang_0.4.11
## [37] systemfonts_1.0.2 splines_4.1.3 broom_0.7.9
## [40] checkmate_2.0.0 inline_0.3.19 yaml_2.2.1
## [43] reshape2_1.4.4 abind_1.4-5 modelr_0.1.8
## [46] threejs_0.3.3 crosstalk_1.1.1 backports_1.2.1
## [49] httpuv_1.6.3 rsconnect_0.8.24 Hmisc_4.6-0
## [52] gridtext_0.1.4 tensorA_0.36.2 tools_4.1.3
## [55] bookdown_0.24 ellipsis_0.3.2 RColorBrewer_1.1-2
## [58] jquerylib_0.1.4 posterior_1.1.0 ggridges_0.5.3
## [61] plyr_1.8.6 base64enc_0.1-3 ps_1.6.0
## [64] prettyunits_1.1.1 rpart_4.1-15 zoo_1.8-9
## [67] cluster_2.1.2 haven_2.4.3 fs_1.5.0
## [70] magrittr_2.0.1 data.table_1.14.2 colourpicker_1.1.1
## [73] reprex_2.0.1 tmvnsim_1.0-2 mvtnorm_1.1-2
## [76] matrixStats_0.61.0 stringfish_0.15.2 qs_0.25.1
## [79] hms_1.1.1 shinyjs_2.0.0 mime_0.12
## [82] evaluate_0.14 arrayhelpers_1.1-0 xtable_1.8-4
## [85] shinystan_2.5.0 jpeg_0.1-9 readxl_1.3.1
## [88] gridExtra_2.3 rstantools_2.1.1 compiler_4.1.3
## [91] V8_3.4.2 crayon_1.4.1 minqa_1.2.4
## [94] StanHeaders_2.21.0-7 htmltools_0.5.2 mgcv_1.8-38
## [97] later_1.3.0 tzdb_0.1.2 Formula_1.2-4
## [100] RcppParallel_5.1.4 RApiSerialize_0.1.0 lubridate_1.8.0
## [103] DBI_1.1.1 dbplyr_2.1.1 MASS_7.3-54
## [106] boot_1.3-28 cli_3.0.1 parallel_4.1.3
## [109] insight_0.14.4 igraph_1.2.6 pkgconfig_2.0.3
## [112] foreign_0.8-81 xml2_1.3.2 svUnit_1.0.6
## [115] dygraphs_1.1.1.6 pbivnorm_0.6.0 svglite_2.0.0
## [118] bslib_0.3.1 webshot_0.5.2 estimability_1.3
## [121] rvest_1.0.1 snakecase_0.11.0 distributional_0.2.2
## [124] callr_3.7.0 digest_0.6.28 rmarkdown_2.11
## [127] cellranger_1.1.0 htmlTable_2.2.1 curl_4.3.2
## [130] shiny_1.7.1 gtools_3.9.2 nloptr_1.2.2.2
## [133] lifecycle_1.0.1 nlme_3.1-153 jsonlite_1.7.2
## [136] viridisLite_0.4.0 fansi_0.5.0 pillar_1.6.3
## [139] lattice_0.20-45 loo_2.4.1 survival_3.2-13
## [142] fastmap_1.1.0 httr_1.4.2 pkgbuild_1.2.0
## [145] glue_1.4.2 xts_0.12.1 png_0.1-7
## [148] shinythemes_1.2.0 stringi_1.7.5 sass_0.4.0
## [151] textshaping_0.3.5 latticeExtra_0.6-29 renv_0.14.0